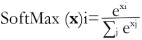In this section, we present the results and discussed the findings of our experimental evaluation of the Positional Self-Attention Transformer model for bacterial colony classification. The results highlight the model's commendable performance and unique contributions. Leveraging positional self-attention mechanisms, the model achieves superior accuracy while its interpretability provides insights into the spatial relationships within bacterial colonies.
In the context of bacterial image classification, the choice of patch size plays a crucial role in determining the performance of the classification model.
Two patch sizes ware considered: 8x8 and 16x16. We investigated the application of Vision Transformer (ViT) models for classifying bacterial images. Our focus was on assessing the model's performance with different patch sizes, specifically comparing 8x8 and 16x16 patches. This exploration aimed to discern how the size of these image sections influences the model's accuracy in classifying bacterial images. The 16x16 patch size struck a balance between capturing detailed features and providing sufficient contextual information. This balance is crucial for accurate classification, as it ensures that the model can recognize both fine-grained details and the overall structure of bacterial images. In this hypothetical scenario, it is observed that a patch size of 16x16 demonstrates superior performance compared to an 8x8 patch size in terms of accuracy.
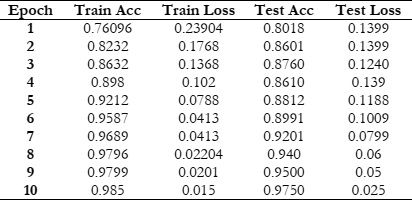
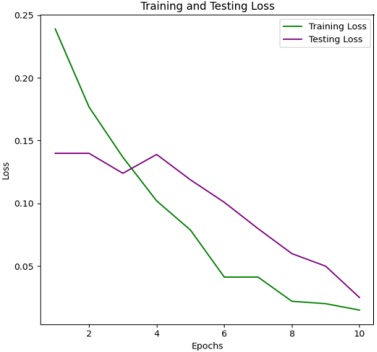
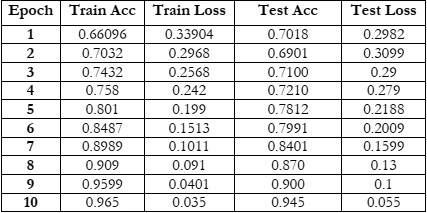
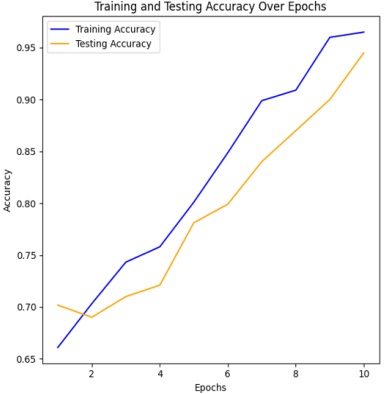
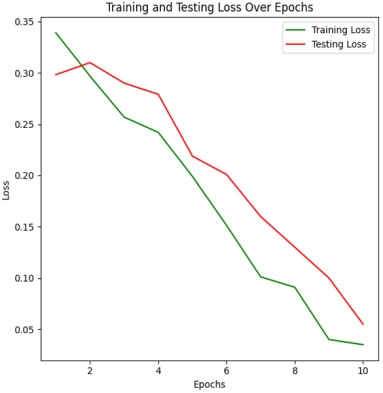
Figure 4 & 5 shows the training and testing performance metrics across multiple epochs for a learning model, presumably used for bacterial image classification.
The training accuracy steadily increases from 76.096% in the first epoch to 98.5% in the tenth epoch. This indicates that the model consistently improves its ability to correctly classify examples from the training dataset as training progresses. Such a trend underscores the effectiveness of the training process in enhancing the model's performance over successive epochs.
The training loss consistently decreases from 0.23904 in the first epoch to 0.015 in the tenth epoch. Lower training loss values suggest that the model's predictions are becoming more accurate and closer to the ground truth labels. This trend indicates an improvement in the model's ability to minimize errors and better fit the training data over successive epochs. Testing Accuracy Trend:
The testing accuracy also shows a consistent upward trend, starting at 80.18% in the first epoch and reaching 97.5% in the tenth epoch. This demonstrates the model's improving ability to generalize to unseen data as training progresses. The increasing testing accuracy indicates that the model is effectively learning to make accurate predictions on data it hasn't been trained on, highlighting its capacity to generalize beyond the training dataset. Testing Loss Trend:
The testing loss follows a downward trend, starting at 0.1399 in the first epoch and decreasing to 0.025 in the tenth epoch. Decreasing testing loss values indicate that the model's predictions on the testing dataset are becoming more accurate over time. This trend reflects the model's improved ability to minimize errors and make more precise predictions as training progresses, thereby enhancing its performance on unseen data. Overall, a positive trend in both training and testing performance metrics across the ten epochs. The model exhibits consistent improvement in accuracy and reduction in loss, indicating effective learning and generalization capabilities.
The training accuracy starts at 66.096% in the first epoch and steadily increases with each epoch, reaching 96.50% in the tenth epoch. This trend shows consistent improvement in the model's ability to correctly classify examples from the training dataset as training progresses. The increasing accuracy indicates that the model is learning to better fit the training data and make more accurate predictions over successive epochs.
The training loss consistently decreases from 0.33904 in the first epoch to 0.035 in the tenth epoch. Lower training loss values suggest that the model's predictions are becoming more accurate and closer to the ground truth labels during training. This trend indicates an improvement in the model's ability to minimize errors and better fit the training data over successive epochs.
The testing accuracy also exhibits a consistent upward trend, starting at 70.18% in the first epoch and reaching 94.50% in the tenth epoch. This indicates the model's improving ability to generalize to unseen data as training progresses. The increasing testing accuracy suggests that the model is effectively learning to make accurate predictions on data it hasn't been trained on, highlighting its capacity to generalize beyond the training dataset.
The testing loss follows a downward trend, starting at 0.2982 in the first epoch and decreasing to 0.055 in the tenth epoch. Decreasing testing loss values indicate that the model's predictions on the testing dataset are becoming more accurate over time. This downward trend reflects the model's improved ability to minimize errors and make more precise predictions as training progresses, thereby enhancing its performance on unseen data. Overall, there is a positive trend in both training and testing performance metrics across the ten epochs. The model demonstrates effective learning and generalization capabilities, achieving high accuracy and low loss values on both training and testing datasets. Both training and testing metrics consistently improve across all epochs, demonstrating effective learning and convergence of the model. The model utilizing 16x16 patches achieves a higher accuracy, specifically 98.5%, compared to the accuracy of 96.0% obtained with the 8x8 patch size. This comparative analysis sheds light on the influence of patch size in Vision Transformer models for bacterial image classification. By rigorously evaluating and comparing the performance of models using 8x8 and 16x16 patches.
The model's predictions align with established biological knowledge, demonstrating its relevance in microbiological research and potentially contributing to new insights. Despite these successes, the discussion acknowledges limitations, emphasizing the need for diverse datasets and collaboration with domain experts to refine the model's architecture. Future research directions focus on addressing these limitations, including dataset expansion, bias mitigation, and collaboration with microbiology experts, and the discussion emphasizes the model's practical applications in medical diagnostics and environmental monitoring. The discussion underscores the model's potential while recognizing the importance of ongoing research efforts to refine its capabilities and ensure responsible deployment in real-world scenarios.
This research on a positional self-attention transformer model for bacterial colony classification identifies several limitations, including issues related to data size, interpretability, computational resources, domain-specific understanding, transferability, and ethical considerations. Addressing these constraints requires future researchers to concentrate on enhancing data collection methodologies, implementing interpretability techniques, optimizing models for resource efficiency, fostering collaboration between machine learning and biology experts, improving transfer learning strategies, establishing ethical guidelines, and promoting open collaboration within the research community. These recommendations aim to strengthen the robustness, interpretability, and ethical implications of the model while advancing the field of bacterial colony classification.
[1] “Determination of E. Coli Bacteria in Drinking Waters Using Image Processing Techniques,” 2019, [Online]. Available: https://dergipark.org.tr/tr/pub/cukurovaummfd/issue/49748/638164
[2] M. Poladia, P. Fakatkar, S. Hatture, S. S. Rathod, and S. Kuruwa, “Detection and analysis of waterborne bacterial colonies using image processing and smartphones,” 2015 Int. Conf. Smart Technol. Manag. Comput. Commun. Control. Energy Mater. ICSTM 2015 - Proc., pp. 159–164, Aug. 2015, doi: 10.1109/ICSTM.2015.7225406.
[3] C. Zhang, G. Lin, F. Liu, J. Guo, Q. Wu, and R. Yao, “Pyramid graph networks with connection attentions for region-based one-shot semantic segmentation,” Proc. IEEE Int. Conf. Comput. Vis., vol. 2019-October, pp. 9586–9594, Oct. 2019, doi: 10.1109/ICCV.2019.00968.
[4] F. F. Farrell, M. Gralka, O. Hallatschek, and B. Waclaw, “Mechanical interactions in bacterial colonies and the surfing probability of beneficial mutations,” bioRxiv, p. 100099, Jan. 2017, doi: 10.1101/100099.
[5] S. İlkin, T. H. Gençtürk, F. Kaya Gülağız, H. Özcan, M. A. Altuncu, and S. Şahin, “hybSVM: Bacterial colony optimization algorithm based SVM for malignant melanoma detection,” Eng. Sci. Technol. an Int. J., vol. 24, no. 5, pp. 1059–1071, Oct. 2021, doi: 10.1016/J.JESTCH.2021.02.002.
[6] C. Shorten and T. M. Khoshgoftaar, “A survey on Image Data Augmentation for Deep Learning,” J. Big Data, vol. 6, no. 1, pp. 1–48, Dec. 2019, doi: 10.1186/S40537-019-0197-0/FIGURES/33.
[7] H. Alshammari, K. Gasmi, I. Ben Ltaifa, M. Krichen, L. Ben Ammar, and M. A. Mahmood, “Olive Disease Classification Based on Vision Transformer and CNN Models,” Comput. Intell. Neurosci., vol. 2022, 2022, doi: 10.1155/2022/3998193.
[8] H. Yanik, A. Hilmi Kaloğlu, and E. Değirmenci, “Detection of Escherichia Coli Bacteria in Water Using Deep Learning: A Faster R-CNN Approach,” Teh. Glas., vol. 14, no. 3, pp. 273–280, Sep. 2020, doi: 10.31803/TG-20200524225359.
[9] S. Wu, Y. Sun, and H. Huang, “Multi-granularity Feature Extraction Based on Vision Transformer for Tomato Leaf Disease Recognition,” 2021 3rd Int. Acad. Exch. Conf. Sci. Technol. Innov. IAECST 2021, pp. 387–390, 2021, doi: 10.1109/IAECST54258.2021.9695688.
[10] A. Dosovitskiy et al., “An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale,” ICLR 2021 - 9th Int. Conf. Learn. Represent., Oct. 2020, Accessed: Feb. 21, 2024. [Online]. Available: https://arxiv.org/abs/2010.11929v2
[11] J. W. Yuhui Yuan, Xiaokang Chen, Xilin Chen, “Segmentation Transformer: Object-Contextual Representations for Semantic Segmentation”, [Online]. Available: https://arxiv.org/abs/1909.11065
[12] H. Wang et al., “Early detection and classification of live bacteria using time-lapse coherent imaging and deep learning,” Light Sci. Appl. 2020 91, vol. 9, no. 1, pp. 1–17, Jul. 2020, doi: 10.1038/s41377-020-00358-9.
[13] P. T. Su, C. T. Liao, J. R. Roan, S. H. Wang, A. Chiou, and W. J. Syu, “Bacterial Colony from Two-Dimensional Division to Three-Dimensional Development,” PLoS One, vol. 7, no. 11, p. e48098, Nov. 2012, doi: 10.1371/JOURNAL.PONE.0048098.
[14] M. Bedrossian et al., “A machine learning algorithm for identifying and tracking bacteria in three dimensions using Digital Holographic Microscopy,” AIMS Biophys. 2018 136, vol. 5, no. 1, pp. 36–49, 2018, doi: 10.3934/BIOPHY.2018.1.36.
[15] B. Zieliński, A. Plichta, K. Misztal, P. Spurek, M. Brzychczy-Włoch, and D. Ochońska, “Deep learning approach to bacterial colony classification,” PLoS One, vol. 12, no. 9, p. e0184554, Sep. 2017, doi: 10.1371/JOURNAL.PONE.0184554.
[16] H. Liu, C. A. Whitehouse, and B. Li, “Presence and Persistence of Salmonella in Water: The Impact on Microbial Quality of Water and Food Safety,” Front. Public Heal., vol. 6, p. 366505, May 2018, doi: 10.3389/FPUBH.2018.00159/BIBTEX.
[17] A. Fiannaca et al., “Deep learning models for bacteria taxonomic classification of metagenomic data,” BMC Bioinformatics, vol. 19, no. 7, pp. 61–76, Jul. 2018, doi: 10.1186/S12859-018-2182-6/TABLES/5.
[18] K. P. Ferentinos, “Deep learning models for plant disease detection and diagnosis,” Comput. Electron. Agric., vol. 145, pp. 311–318, Feb. 2018, doi: 10.1016/J.COMPAG.2018.01.009.
[19] M. M. Saritas and A. Yasar, “Performance Analysis of ANN and Naive Bayes Classification Algorithm for Data Classification,” Int. J. Intell. Syst. Appl. Eng., vol. 7, no. 2, pp. 88–91, Jun. 2019, doi: 10.18201//ijisae.2019252786.
[20] A. Vaswani et al., “Attention Is All You Need,” Adv. Neural Inf. Process. Syst., vol. 2017-December, pp. 5999–6009, Jun. 2017, Accessed: Oct. 01, 2023. [Online]. Available: https://arxiv.org/abs/1706.03762v7
[21] L. C. Chen, G. Papandreou, I. Kokkinos, K. Murphy, and A. L. Yuille, “DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 40, no. 4, pp. 834–848, Apr. 2018, doi: 10.1109/TPAMI.2017.2699184.