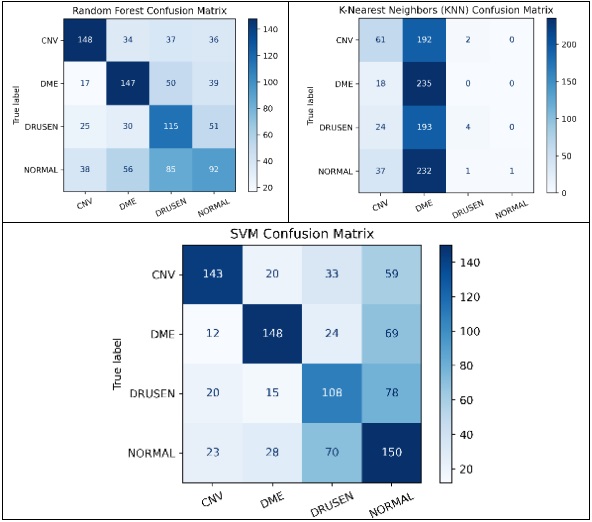
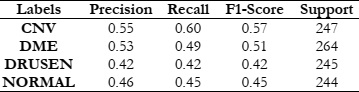
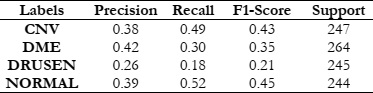
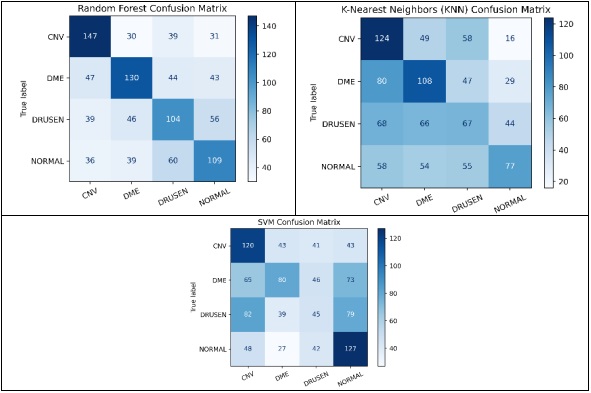
Our experiments revealed promising results across different feature extraction methods and machine learning classifiers. Notably, the use of a HOG in conjunction with a SVM classifier outperformed other combinations, achieving a maximum accuracy of 78.8%. This indicates the effectiveness of HOG in capturing discriminative features from retinal OCT images, facilitating accurate disease classification.
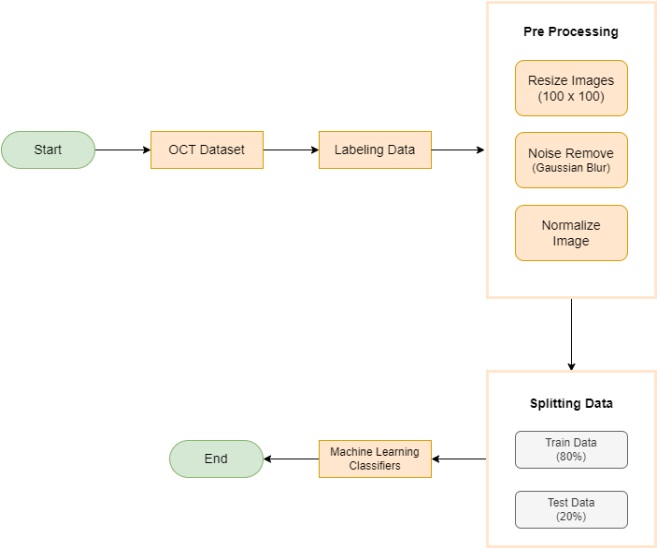
The study offers a detailed comparison across various models for retinal disease distribution, emphasizing both time efficiency and accuracy tailored to the specific requirements of each method. We have employed three distinct feature extraction techniques in this research: HOG, LBP, and FOSF. Additionally, the study incorporates three widely used classification approaches: RFC, SVM, and KNN.
The HOG is a feature descriptor extensively used in computer vision and image processing for object detection tasks. It operates by segmenting an image into smaller, overlapping sections, calculating the gradients in each section, and then categorizing these gradient orientations into histograms. This process generates feature vectors that effectively encapsulate the local intensity gradients, offering a robust representation of the shapes and structures of objects. In our study, we have employed the HOG feature descriptor for the classification of OCT images, leveraging its renowned capabilities in computer vision and image processing. We applied the HOG algorithm to our dataset, which involved breaking down images into small, overlapping sections, computing the local intensity gradients, and then generating histograms that characterize the gradient orientations.
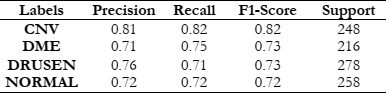
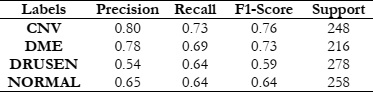
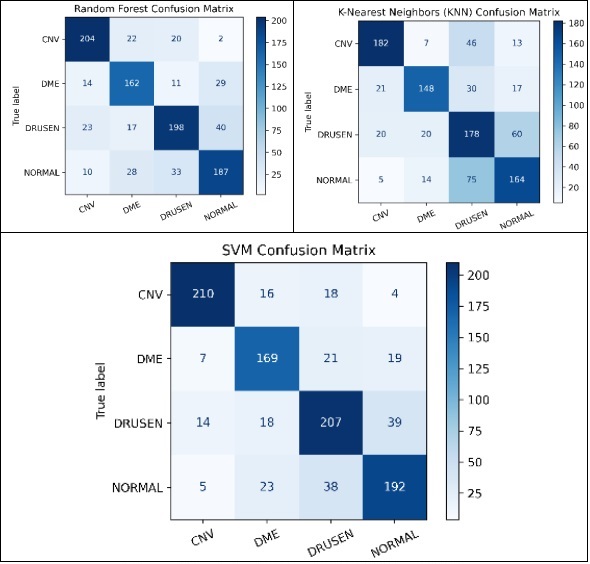
To assess the effectiveness of HOG features in object recognition, we utilized three distinct classifiers: RFC, SVM, and KNN. Our experimental approach included a meticulous division of the dataset into training and test sets, with each classifier configured appropriately. The results were comprehensively analyzed using standard evaluation metrics such as accuracy, precision, recall, and F1-score. This analysis aimed to elucidate the performance of each classifier in accurately distinguishing between different object classes, thereby highlighting the potential of HOG features in object detection and classification.
The LBP is a texture descriptor widely utilized for texture analysis and classification in image processing. It functions by comparing a central pixel's intensity with that of its surrounding pixels, encoding these relational intensities into binary patterns. These patterns are subsequently converted into histograms, effectively encapsulating the texture features of the image. LBP is particularly adept at identifying textural patterns such as edges, corners, and diverse texture variations. In our study, we applied the LBP method to our dataset and engaged three different classifiers for evaluation: RFC, SVM, and KNN. The goal was to determine the effectiveness of LBP, in combination with these classifiers, in differentiating various textures within the dataset, thereby offering valuable insights into its applicability for texture-centric image classification tasks.
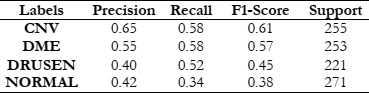
To gauge the efficacy of the LBP when used alongside the RFC, SVM, and KNN classifiers, our evaluation focused on key metrics such as accuracy, recall, precision, and the F1 score. These metrics collectively serve as critical indicators of the classifiers' proficiency in accurately detecting and classifying different textures present in the images. Accuracy indicates the overall correctness of the classifications made, recall measures the ability of the classifiers to correctly identify relevant examples of each texture class, precision assesses the exactness of the classifiers in correctly labeling instances of a specific class, and the F1 score provides a harmonized evaluation, considering both precision and recall. These performance indicators present a holistic view of the effectiveness of LBP, in synergy with RFC, SVM, and KNN, in precisely classifying textures within our dataset. This analysis thereby contributes significant insights to the broader domain of texture-based image analysis.
The FOSF is a method that employs the Fourier transform to derive spectral features from signals or images. In image processing, FOSF translates the spatial details of an image into the frequency domain, revealing its frequency components. This conversion is pivotal in various tasks, including image compression, filtering, and feature extraction. FOSF is especially useful in fields where analyzing an image's frequency content is essential, such as in medical or satellite image analysis. To examine its effectiveness, we applied FOSF to our dataset and engaged three classifiers for assessment: RFC, SVM and KNN. This technique is particularly advantageous in situations where discerning an image's frequency details is crucial, for instance in medical imaging or satellite imagery analysis.
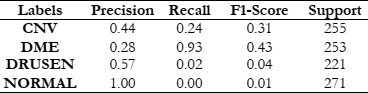
The next phase of our study focused on evaluating how FOSF, combined with these classifiers, performs in image processing tasks like classification and object detection. We assessed the performance using key metrics such as accuracy, recall, precision, and F1 score. This comprehensive analysis was aimed at determining the proficiency of FOSF, in conjunction with Random Forest, SVM, and KNN, in accurately differentiating various patterns and structures within our dataset. The results provided a nuanced understanding of FOSF's capabilities when integrated with different classification methods, offering valuable insights into its potential applications in the field of image analysis.
The results of this study provide fascinating insights into the relative effectiveness of the explored methods. Universally, the SVM demonstrated superior performance compared to the RFC and KNN, excelling not only in accuracy but also in computational efficiency.
This paper goes beyond merely comparing different feature extraction techniques and classifiers for Retinal Disease detection; it also emphasizes the importance of judiciously choosing the most effective combination of filters and classifiers to achieve enhanced accuracy. The results underscore the notable benefits of integrating HOG with the SVM Classifier. This provides critical insights for future studies and practical implementations in medical image analysis, highlighting a promising direction for advancements in this field.
[1] J. Jiang et al., “Ultrahigh speed Spectral / Fourier domain OCT ophthalmic imaging at 70,000 to 312,500 axial scans per second,” Opt. Express, Vol. 16, Issue 19, pp. 15149-15169, vol. 16, no. 19, pp. 15149–15169, Sep. 2008, doi: 10.1364/OE.16.015149.
[2] M. E. J. van Velthoven, D. J. Faber, F. D. Verbraak, T. G. van Leeuwen, and M. D. de Smet, “Recent developments in optical coherence tomography for imaging the retina,” Prog. Retin. Eye Res., vol. 26, no. 1, pp. 57–77, Jan. 2007, doi: 10.1016/J.PRETEYERES.2006.10.002.
[3] S. A. Boppart, G. J. Tearney, B. E. Bouma, J. F. Southern, M. E. Brezinski, and J. G. Fujimoto, “Noninvasive assessment of the developing Xenopus cardiovascular system using optical coherence tomography,” Proc. Natl. Acad. Sci. U. S. A., vol. 94, no. 9, pp. 4256–4261, Apr. 1997, doi: 10.1073/PNAS.94.9.4256/ASSET/4849CBA2-F776-442F-86D0-A06D9DB929CE/ASSETS/GRAPHIC/PQ0970492007.JPEG.
[4] M. J. Suter et al., “Intravascular Optical Imaging Technology for Investigating the Coronary Artery,” JACC Cardiovasc. Imaging, vol. 4, no. 9, pp. 1022–1039, Sep. 2011, doi: 10.1016/J.JCMG.2011.03.020.
[5] J. F. Southern et al., “Scanning single-mode fiber optic catheter–endoscope for optical coherence tomography,” Opt. Lett. Vol. 21, Issue 7, pp. 543-545, vol. 21, no. 7, pp. 543–545, Apr. 1996, doi: 10.1364/OL.21.000543.
[6] G. J. Tearney et al., “In Vivo Endoscopic Optical Biopsy with Optical Coherence Tomography,” Science (80-. )., vol. 276, no. 5321, pp. 2037–2039, Jun. 1997, doi: 10.1126/SCIENCE.276.5321.2037.
[7] T. Gambichler, G. Moussa, M. Sand, D. Sand, P. Altmeyer, and K. Hoffmann, “Applications of optical coherence tomography in dermatology,” J. Dermatol. Sci., vol. 40, no. 2, pp. 85–94, Nov. 2005, doi: 10.1016/j.jdermsci.2005.07.006.
[8] J. M. Schmitt, M. J. Yadlowsky, and R. F. Bonner, “Subsurface Imaging of Living Skin with Optical Coherence Microscopy,” Dermatology, vol. 191, no. 2, pp. 93–98, Feb. 1995, doi: 10.1159/000246523.
[9] “Non-invasive ophthalmic imaging of adult zebrafish eye using optical coherence tomography.” Accessed: Feb. 13, 2024. [Online]. Available: https://core.ac.uk/download/pdf/291515351.pdf
[10] J. S. Schuman, L. Kagemann, H. Ishikawa, and G. Wollstein, “Spectral-Domain Optical Coherence Tomography as a Noninvasive Method to Assess Damaged and Regenerating Adult Zebrafish Retinas,” Invest. Ophthalmol. Vis. Sci., vol. 53, no. 11, pp. 7315–7315, Oct. 2012, doi: 10.1167/IOVS.12-10925.
[11] “Optical coherence tomography for high-resolution imaging of mouse development in utero.” Accessed: Feb. 13, 2024. [Online]. Available: https://www.spiedigitallibrary.org/journals/journal-of-biomedical-optics/volume-16/issue-04/046004/Optical-coherence-tomography-for-high-resolution-imaging-of-mouse-development/10.1117/1.3560300.full#_=_
[12] C. A. Stewart, I. V. Larina, J. C. Burton, S. Wang, and R. R. Behringer, “High-resolution three-dimensional in vivo imaging of mouse oviduct using optical coherence tomography,” Biomed. Opt. Express, Vol. 6, Issue 7, pp. 2713-2723, vol. 6, no. 7, pp. 2713–2723, Jul. 2015, doi: 10.1364/BOE.6.002713.
[13] A. Alex et al., “A Circadian Clock Gene, Cry, Affects Heart Morphogenesis and Function in Drosophila as Revealed by Optical Coherence Microscopy,” PLoS One, vol. 10, no. 9, p. e0137236, Sep. 2015, doi: 10.1371/JOURNAL.PONE.0137236.
[14] M. Watanabe et al., “Ultrahigh-speed optical coherence tomography imaging and visualization of the embryonic avian heart using a buffered Fourier Domain Mode Locked laser,” Opt. Express, Vol. 15, Issue 10, pp. 6251-6267, vol. 15, no. 10, pp. 6251–6267, May 2007, doi: 10.1364/OE.15.006251.
[15] F. Shi et al., “Automated 3-D retinal layer segmentation of macular optical coherence tomography images with serous pigment epithelial detachments,” IEEE Trans. Med. Imaging, vol. 34, no. 2, pp. 441–452, Feb. 2015, doi: 10.1109/TMI.2014.2359980.
[16] J. Sugmk, S. Kiattisin, and A. Leelasantitham, “Automated classification between age-related macular degeneration and Diabetic macular edema in OCT image using image segmentation,” BMEiCON 2014 - 7th Biomed. Eng. Int. Conf., Jan. 2014, doi: 10.1109/BMEICON.2014.7017441.
[17] A. Lang, A. Carass, B. M. Jedynak, S. D. Solomon, P. A. Calabresi, and J. L. Prince, “Intensity inhomogeneity correction of macular OCT using N3 and retinal flatspace,” Proc. - Int. Symp. Biomed. Imaging, vol. 2016-June, pp. 197–200, Jun. 2016, doi: 10.1109/ISBI.2016.7493243.
[18] M. Adhi et al., “Choroidal analysis in healthy eyes using swept-source optical coherence tomography compared to spectral domain optical coherence tomography,” Am. J. Ophthalmol., vol. 157, no. 6, pp. 1272-1281.e1, Jun. 2014, doi: 10.1016/j.ajo.2014.02.034.
[19] M. A. Hussain et al., “Classification of healthy and diseased retina using SD-OCT imaging and Random Forest algorithm,” PLoS One, vol. 13, no. 6, p. e0198281, Jun. 2018, doi: 10.1371/JOURNAL.PONE.0198281.
[20] M. Wojtkowski et al., “Ophthalmic imaging by spectral optical coherence tomography,” Am. J. Ophthalmol., vol. 138, no. 3, pp. 412–419, Sep. 2004, doi: 10.1016/j.ajo.2004.04.049.
[21] D. S. Ting, L. R. Pasquale, L. Peng, J. P. Campbell, A. Y. Lee, R. Raman, G. S. Tan, L. Schmetterer, P. A. Keane, and T. Y. Wong, “Artificial intelligence and deep learning in ophthalmology,” British Journal of Ophthalmology, vol. 103, pp. 167–175, 2019.
[22] Schmidt-Erfurth, et al, “Unsupervised identification of disease marker candidates in retinal oct imaging data,” IEEE transactions on medical imaging, vol. 38, pp. 1037–1047, 2019.
[23] Lee C.S., Baughman D.M., Lee A.Y. Deep Learning Is Effective for Classifying Normal versus Age-Related Macular Degeneration OCT Images. Ophthalmol. Retin. 2017;1:322–327. doi: 10.1016/j.oret.2016.12.009.
[24] Kermany DS, Goldbaum M, Cai W, Lewis MA. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning Resource Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell. 2018;172:1122–31.e9. https://doi.org/10.1016/j.cell.2018.02.010.
[25] Huang L., He X., Fang L., Rabbani H., Chen X. Automatic Classification of Retinal Optical Coherence Tomography Images With Layer Guided Convolutional Neural Network. IEEE Signal Process. Lett. 2019;26:1026–1030. doi: 10.1109/LSP.2019.2917779.
[26] Chowdhary C.L., Acharjya D. Clustering Algorithm in Possibilistic Exponential Fuzzy C-Mean Segmenting Medical Images. J. Biomimetics Biomater. Biomed. Eng. 2017;30:12–23. doi: 10.4028/www.scientific.net/JBBBE.30.12.
[27] T. Tsuji et al., “Classification of optical coherence tomography images using a capsule network,” BMC Ophthalmol., vol. 20, no. 1, pp. 1–9, Mar. 2020, doi: 10.1186/S12886-020-01382-4/FIGURES/9.
[28] G. Latha and P. Aruna Priya, “Glaucoma Retinal Image Detection and Classification using Machine Learning Algorithms,” J. Phys. Conf. Ser., vol. 2335, no. 1, p. 012025, Sep. 2022, doi: 10.1088/1742-6596/2335/1/012025.
[29] Y. Zhou, “Automated Identification of Diabetic Retinopathy Using Deep Learning,” 2021.
[30] Jian Li, “Automated Detection and Classification of Diabetic Retinopathy Using Deep Learning Based on EfficientNet,” 2020.
[31] H. Fu, “Automated diagnosis of diabetic retinopathy using deep learning”, 2018
[32] S. W. Ting et al., “Artificial intelligence and deep learning in ophthalmology,” Br. J. Ophthalmol., vol. 103, no. 2, pp. 167–175, Feb. 2019, doi: 10.1136/BJOPHTHALMOL-2018-313173.
[33] R. Gargeya and T. Leng, “Automated Identification of Diabetic Retinopathy Using Deep Learning,” Ophthalmology, vol. 124, no. 7, pp. 962–969, Jul. 2017, doi: 10.1016/J.OPHTHA.2017.02.008.
[34] Hwang D.K., Hsu C.C., Chang K.J., Chao D., Sun C.H., Jheng Y.C., Yarmishyn A.A., Wu J.C., Tsai C.Y., Wang M.L., et al. Artificial intelligence-based decision-making for age-related macular degeneration. Theranostics. 2019;9:232–245. doi: 10.7150/thno.28447.
[35] Tasnim N., Hasan M., Islam I. Comparisonal study of Deep Learning approaches on Retinal OCT Image. arXiv. 20191912.07783