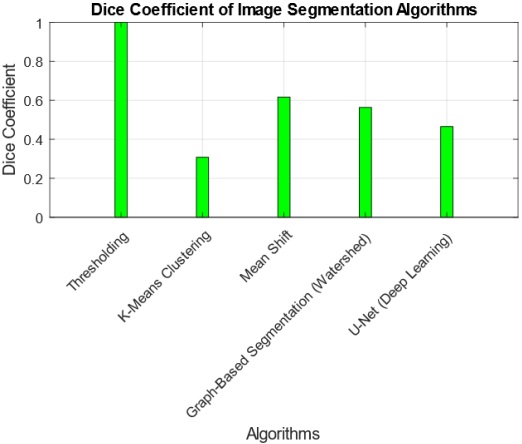
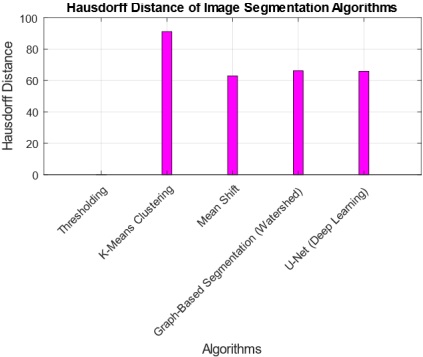
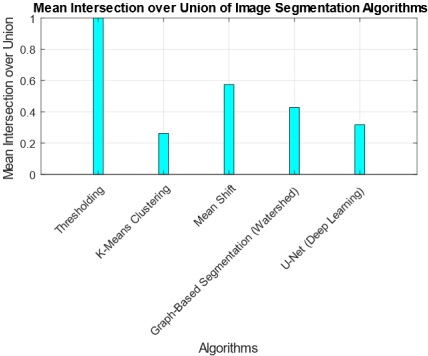
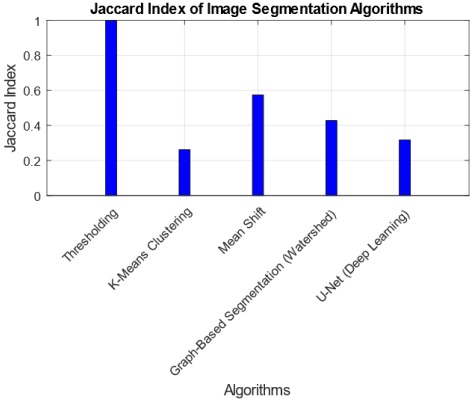
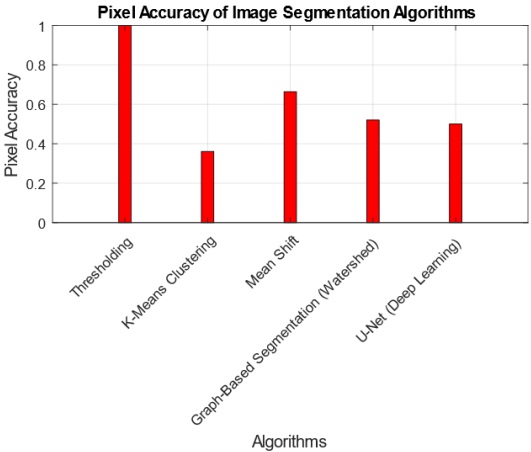
The interpretation of findings from the comparative analysis of five image segmentation algorithms provides valuable insights into their performance across multiple metrics. The Threshold Algorithm consistently outperforms its counterparts, achieving perfect scores in Jaccard Index, Dice Coefficient, Pixel Accuracy, Haudorff Distance, and Mean Intersection over Union. This suggests that the Threshold Algorithm excels in accurately delineating segmentation boundaries and achieving high pixel-level accuracy. Practical implications of these findings indicate that the Threshold Algorithm is a robust choice for applications demanding precise image segmentation, such as medical imaging or object detection. However, the research also highlights tradeoffs, as some algorithms, like K-Means and U-Net, exhibit lower scores in certain metrics, emphasizing the need for careful consideration of specific requirements in choosing an algorithm. The tradeoff between complexity and accuracy is evident, with more complex algorithms potentially introducing challenges in certain scenarios. Overall, these findings contribute to a nuanced understanding of algorithmic performance, guiding practitioners in selecting the most suitable segmentation approach based on their application-specific needs.
[1] Z. Wang, E. Wang, and Y. Zhu, “Image segmentation evaluation: a survey of methods,” Artif. Intell. Rev., vol. 53, no. 8, pp. 5637–5674, Dec. 2020, doi: 10.1007/S10462-020-09830-9/METRICS.
[2] P. Jaglan, R. Dass, and M. Duhan, “A Comparative Analysis of Various Image Segmentation Techniques,” Lect. Notes Networks Syst., vol. 46, pp. 359–374, 2019, doi: 10.1007/978-981-13-1217-5_36/COVER.
[3] “A survey on segmentation techniques for image processing.” Accessed: Feb. 26, 2024. [Online]. Available: https://ijeee.edu.iq/Papers/Vol17-Issue2/1570736047.pdf
[4] S. Singh, N. Mittal, D. Thakur, H. Singh, D. Oliva, and A. Demin, “Nature and Biologically Inspired Image Segmentation Techniques,” Arch. Comput. Methods Eng. 2021 293, vol. 29, no. 3, pp. 1415–1442, Jun. 2021, doi: 10.1007/S11831-021-09619-1.
[5] S. Song and T. Gao, “Research on image segmentation algorithm based on threshold,” Proc. - 2021 13th Int. Conf. Meas. Technol. Mechatronics Autom. ICMTMA 2021, pp. 306–308, Jan. 2021, doi: 10.1109/ICMTMA52658.2021.00071.
[6] K. K. D. Ramesh, G. Kiran Kumar, K. Swapna, D. Datta, and S. Suman Rajest, “A Review of Medical Image Segmentation Algorithms,” EAI Endorsed Trans. Pervasive Heal. Technol., vol. “7,” no. 27, Apr. 2021, doi: 10.4108/EAI.12-4-2021.169184.
[7] S. Pare, A. Kumar, G. K. Singh, and V. Bajaj, “Image Segmentation Using Multilevel Thresholding: A Research Review,” Iran. J. Sci. Technol. - Trans. Electr. Eng., vol. 44, no. 1, pp. 1–29, Mar. 2020, doi: 10.1007/S40998-019-00251-1/METRICS.
[8] M. Abdel-Basset, V. Chang, and R. Mohamed, “A novel equilibrium optimization algorithm for multi-thresholding image segmentation problems,” Neural Comput. Appl., vol. 33, no. 17, pp. 10685–10718, Sep. 2021, doi: 10.1007/S00521-020-04820-Y/METRICS.
[9] C. Shi, Y. Zhou, B. Qiu, D. Guo, and M. Li, “CloudU-Net: A Deep Convolutional Neural Network Architecture for Daytime and Nighttime Cloud Images’ Segmentation,” IEEE Geosci. Remote Sens. Lett., vol. 18, no. 10, pp. 1688–1692, Oct. 2021, doi: 10.1109/LGRS.2020.3009227.
[10] H. S. Lee et al., “Efficient Defect Identification via Oxide Memristive Crossbar Array Based Morphological Image Processing,” Adv. Intell. Syst., vol. 3, no. 2, p. 2000202, Feb. 2021, doi: 10.1002/AISY.202000202.
[11] M. Ahmed, R. Seraj, and S. M. S. Islam, “The k-means Algorithm: A Comprehensive Survey and Performance Evaluation,” Electron. 2020, Vol. 9, Page 1295, vol. 9, no. 8, p. 1295, Aug. 2020, doi: 10.3390/ELECTRONICS9081295.
[12] K. Tian, J. Li, J. Zeng, A. Evans, and L. Zhang, “Segmentation of tomato leaf images based on adaptive clustering number of K-means algorithm,” Comput. Electron. Agric., vol. 165, p. 104962, Oct. 2019, doi: 10.1016/J.COMPAG.2019.104962.
[13] A. M. Ikotun, A. E. Ezugwu, L. Abualigah, B. Abuhaija, and J. Heming, “K-means clustering algorithms: A comprehensive review, variants analysis, and advances in the era of big data,” Inf. Sci. (Ny)., vol. 622, pp. 178–210, Apr. 2023, doi: 10.1016/J.INS.2022.11.139.
[14] T. G. Debelee, F. Schwenker, S. Rahimeto, and D. Yohannes, “Evaluation of modified adaptive k-means segmentation algorithm,” Comput. Vis. Media, vol. 5, no. 4, pp. 347–361, Dec. 2019, doi: 10.1007/S41095-019-0151-2/METRICS.
[15] M. S. Yang and K. P. Sinaga, “A feature-reduction multi-view k-means clustering algorithm,” IEEE Access, vol. 7, pp. 114472–114486, 2019, doi: 10.1109/ACCESS.2019.2934179.
[16] M. Zhao et al., “Faster Mean-shift: GPU-accelerated clustering for cosine embedding-based cell segmentation and tracking,” Med. Image Anal., vol. 71, p. 102048, Jul. 2021, doi: 10.1016/J.MEDIA.2021.102048.
[17] S. Yun and S. Kim, “Robust infrared target tracking using thermal information in mean-shift,” https://doi.org/10.1117/12.2519191, vol. 10995, pp. 52–57, May 2019, doi: 10.1117/12.2519191.
[18] J. W. Hsieh, C. H. Chou, M. C. Chang, P. Y. Chen, S. Santra, and C. S. Huang, “Mean-Shift Based Differentiable Architecture Search,” IEEE Trans. Artif. Intell., 2023, doi: 10.1109/TAI.2023.3329792.
[19] J. J. Hwang et al., “SegSort: Segmentation by discriminative sorting of segments,” Proc. IEEE Int. Conf. Comput. Vis., vol. 2019-October, pp. 7333–7343, Oct. 2019, doi: 10.1109/ICCV.2019.00743.
[20] Q. Huang, Y. Huang, Y. Luo, F. Yuan, and X. Li, “Segmentation of breast ultrasound image with semantic classification of superpixels,” Med. Image Anal., vol. 61, p. 101657, Apr. 2020, doi: 10.1016/J.MEDIA.2020.101657.
[21] X. Jia, T. Lei, P. Liu, D. Xue, H. Meng, and A. K. Nandi, “Fast and Automatic Image Segmentation Using Superpixel-Based Graph Clustering,” IEEE Access, 2020, doi: 10.1109/ACCESS.2020.3039742.
[22] A. Ibrahim, “Image Segmentation Methods Based on Superpixel Techniques: A Survey.” Jan. 01, 2020. Accessed: Feb. 26, 2024. [Online]. Available: https://www.academia.edu/70227250/Image_Segmentation_Methods_Based_on_Superpixel_Techniques_A_Survey
[23] Y. Mourchid, M. El Hassouni, and H. Cherifi, “A general framework for complex network-based image segmentation,” Multimed. Tools Appl., vol. 78, no. 14, pp. 20191–20216, Jul. 2019, doi: 10.1007/S11042-019-7304-2/METRICS.
[24] Y. Zhou, G. Gallego, X. Lu, S. Liu, and S. Shen, “Event-Based Motion Segmentation with Spatio-Temporal Graph Cuts,” IEEE Trans. Neural Networks Learn. Syst., vol. 34, no. 8, pp. 4868–4880, Aug. 2023, doi: 10.1109/TNNLS.2021.3124580.
[25] Y. Zhao et al., “Graph-based region and boundary aggregation for biomedical image segmentation,” IEEE Trans. Med. Imaging, vol. 41, no. 3, pp. 690–701, Mar. 2022, doi: 10.1109/TMI.2021.3123567.
[26] S. Momin, Y. Lei, and Z. Tian, “Medical Image Segmentation Based on U-Net,” J. Phys. Conf. Ser., vol. 2547, no. 1, p. 012010, Jul. 2023, doi: 10.1088/1742-6596/2547/1/012010.
[27] N. Siddique, S. Paheding, C. P. Elkin, and V. Devabhaktuni, “U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications,” IEEE Access, vol. 9, pp. 82031–82057, 2021, doi: 10.1109/ACCESS.2021.3086020.
[28] V. D. Nahian Siddique, Paheding Sidike, Colin Elkin, “U-Net and its variants for medical image segmentation: theory and applications”, [Online]. Available: https://arxiv.org/abs/2011.01118
[29] L. Yin et al., “U-Net-STN: A Novel End-to-End Lake Boundary Prediction Model,” L. 2023, Vol. 12, Page 1602, vol. 12, no. 8, p. 1602, Aug. 2023, doi: 10.3390/LAND12081602.
[30] R. Azad, M. Asadi-Aghbolaghi, M. Fathy, and S. Escalera, “Bi-directional ConvLSTM U-net with densley connected convolutions,” Proc. - 2019 Int. Conf. Comput. Vis. Work. ICCVW 2019, pp. 406–415, Oct. 2019, doi: 10.1109/ICCVW.2019.00052.
[31] “SIPI Image Database - Misc.” Accessed: Dec. 02, 2023. [Online]. Available: https://sipi.usc.edu/database/database.php?volume=misc
[32] T. Eelbode et al., “Optimization for Medical Image Segmentation: Theory and Practice When Evaluating With Dice Score or Jaccard Index,” IEEE Trans. Med. Imaging, vol. 39, no. 11, pp. 3679–3690, Nov. 2020, doi: 10.1109/TMI.2020.3002417.
[33] J. Bertels et al., “Optimizing the Dice Score and Jaccard Index for Medical Image Segmentation: Theory and Practice,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 11765 LNCS, pp. 92–100, 2019, doi: 10.1007/978-3-030-32245-8_11/COVER.
[34] N. A. Muhadi, A. F. Abdullah, S. K. Bejo, M. R. Mahadi, and A. Mijic, “Image Segmentation Methods for Flood Monitoring System,” Water 2020, Vol. 12, Page 1825, vol. 12, no. 6, p. 1825, Jun. 2020, doi: 10.3390/W12061825.
[35] R. R. Shamir, Y. Duchin, J. Kim, G. Sapiro, and N. Harel, “Continuous Dice Coefficient: a Method for Evaluating Probabilistic Segmentations,” bioRxiv, p. 306977, Apr. 2018, doi: 10.1101/306977.
[36] R. Zhao et al., “Rethinking dice loss for medical image segmentation,” Proc. - IEEE Int. Conf. Data Mining, ICDM, vol. 2020-November, pp. 851–860, Nov. 2020, doi: 10.1109/ICDM50108.2020.00094.
[37] B. Cheng, R. Girshick, P. Dollár, A. C. Berg, and A. Kirillov, “Boundary IoU: Improving object-centric image segmentation evaluation,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., pp. 15329–15337, 2021, doi: 10.1109/CVPR46437.2021.01508.
[38] É. O. Rodrigues, “An efficient and locality-oriented Hausdorff distance algorithm: Proposal and analysis of paradigms and implementations,” Pattern Recognit., vol. 117, p. 107989, Sep. 2021, doi: 10.1016/J.PATCOG.2021.107989.
[39] J. T. Jeng, C. M. Chen, S. C. Chang, and C. C. Chuang, “IPFCM Clustering Algorithm Under Euclidean and Hausdorff Distance Measure for Symbolic Interval Data,” Int. J. Fuzzy Syst., vol. 21, no. 7, pp. 2102–2119, Oct. 2019, doi: 10.1007/S40815-019-00707-W/METRICS.
[40] Y.-J. Cho, “Weighted Intersection over Union (wIoU): A New Evaluation Metric for Image Segmentation,” Jul. 2021, Accessed: Feb. 26, 2024. [Online]. Available: https://arxiv.org/abs/2107.09858v4
[41] H. Rezatofighi, N. Tsoi, J. Gwak, A. Sadeghian, I. Reid, and S. Savarese, “Generalized intersection over union: A metric and a loss for bounding box regression,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2019-June, pp. 658–666, Jun. 2019, doi: 10.1109/CVPR.2019.00075.
Appendix: MATLAB Code for Image Segmentation:
Description:
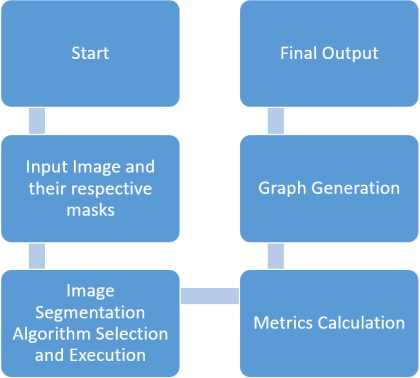
The MATLAB code provided below implements various image segmentation algorithms and evaluates their performance using different metrics such as Jaccard Index, Dice Coefficient, Pixel Accuracy, Hausdorff Distance, and Mean Intersection over Union.
Code Repository Link:
https://www.kaggle.com/datasets/umerijazrandhawa/matlab-code-for-image-segmentation
Code Files:
Main_Script_Segmentation.m:Main script to perform image segmentation and generate evaluation metrics.
Run_Image_Segmentation_and_Metrics.m: Function to run image segmentation for multiple images and calculate evaluation metrics.
Get_Algorithm_Name.m: Function to map algorithm numbers to algorithm names.
Run_Segmentation_Algorithm.m: Function to run specific segmentation algorithms based on algorithm numbers.
Resize_Mask.m: Function to resize masks to a common size.
Algorithm-specific segmentation functions:
thresholding_segmentation.m
kmeans_clustering_segmentation.m
mean_shift_segmentation.m
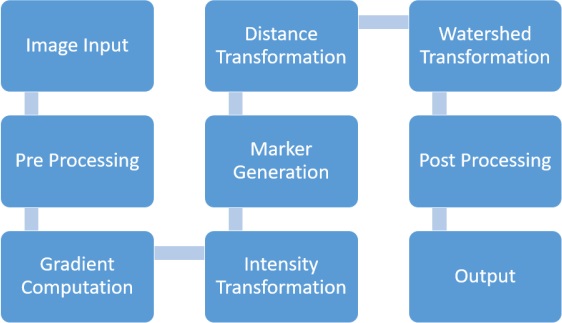
watershed_segmentation.m
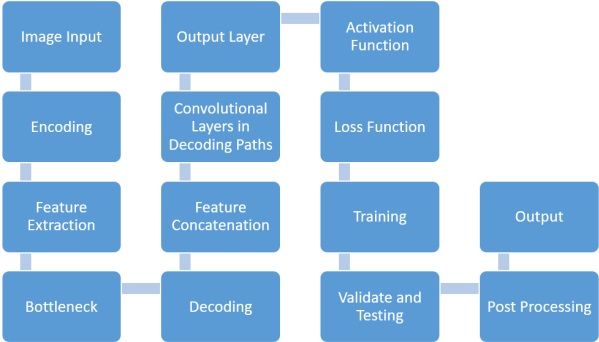
unet_segmentation.m
Evaluation metrics functions:
jaccard_index.m
dice_coefficient.m
pixel_accuracy.m
hausdorff_distance.m
intersection_over_union.m
hausdorff_distance_single.m
kmeans_grayscale.m
mean_shift_grayscale.m
Input Data:
Five sample images (Image1.tiff to Image5.tiff) and their corresponding ground truth masks (mask1.tif to mask5.tif) are used as input data for the image segmentation process.
Output:
The MATLAB code generates graphs illustrating the performance of different segmentation algorithms based on the evaluation metrics mentioned above.
Usage:
• Clone or download the repository containing the MATLAB code.
• Open MATLAB and navigate to the directory containing the downloaded files.
• Run the main_script_segmentation.m script to execute the image segmentation process.