A Deep Learning Framework for Multi-Drug Side Effects Prediction with Drug Chemical Substructure
Muhammad Asad Arshed1,2, Shahzad Mumtaz3, Omer Riaz4, Waqas Sharif5, Saima Abdullah5
1Department of Software Engineering, University of Management & Technology, Lahore, Pakistan
2Department of Artificial Intelligence, The Islamia University of Bahawalpur, Bahawalpur, Pakistan
3Department of Data Science, The Islamia University of Bahawalpur, Bahawalpur, Pakistan
4 Department of Information Technology, The Islamia University of Bahawalpur, Bahawalpur, Pakistan
5Department of Computer Science, The Islamia University of Bahawalpur, Bahawalpur, Pakistan
* Correspondence: Muhammad Asad Arshed – Email ID: muhammadasadarshed@gmail.com
Citation | Arshed. M. A, Mumtaz. S, Riaz. O, Sharif. W and Abdullah. S, “A Deep Learning Framework for Multi-Drug Side Effects Pridiction with Drug Chemical Substructure” International Journal of Innovations in Science and Technology. Vol 4, Issue 1, pp: 19- 31, 2022.
Received | Dec 30, 2021; Revised | Jan 20, 2022 Accepted | Jan 20, 2022; Published | Jan 22, 2022.
________________________________________________________________________
Nowadays, side effects and adverse reactions of drugs are considered the major concern regarding public health. In the process of drug development, it is also considered the main cause of drug failure. Due to the major side effects, drugs are withdrawan from the market immediately. Therefore, in the drug discovery process, the prediction of side effects is a basic need to control the drug development cost and time as well as launching of an effective drug in the market in terms of patient health recovery. In this study, we have proposed a deep learning model named “DLMSE” for the prediction of multiple side effects of drugs with the chemical structure of drugs. As it is a common experience that a single drug can cause multiple side effects, that’s why we have proposed a deep learning model that can predict multiple side effects for a single drug. We have considered three side effects (Dizziness, Allergy, Headache) in this study. We have collected the drug side effects information from the SIDER database. We have achieved an accuracy of ‘0.9494’ with our multi-label classification based proposed model. The proposed model can be used in different stages of the drug development process.
Keywords: Drug Side Effect; Drug Chemical Structure; Deep Learning; Multi-Label Classification; PCA
Introduction
Pharmaceutical companies launch drugs in the market for treatment purposes but most of the drugs are immediately withdrawn from the market (e.g., Rofecoxib) due to side effects and adverse reactions [1]. The average estimated cost to research and develop a drug is $2.6 billion [2]. Therefore, identifying potential side effects is valuable to reduce the risk and cost of drug discovery. Risk can be decreased for the patient as well as pharmaceutical companies with early detection of drug side effects. The drug side effects are considered the 4th largest cause of deaths in the United States that results in one million deaths per year [3]. The failure of drug development is due to the major drug side effects that are unacceptable. Wet experiments are costly and time-consuming in terms of drug discovery. Thus, it needs time to design an effective methods are required, however Clinical trials are expensive and time-consuming. Artificial Intelligence (AI) is also playing an important role in the medical field for the last some years. Due to advancements in AI, several computational methods have come into existence to solve complex medical problems.
In few previous years, several computational models have been proposed for prediction of drug side effects. Some of the proposed model treat side effect as separate classifier [3, 4, 5, 6, 7]. In these studies, samples are consideredpositive if the drugs have a particular side effect, but samples are considered negative if they have no side effects. A large number of classifiers are required to determine the side effects of a particular drug. A single drug can be a cause of multilabel classification problem [8]–[13]. Some models use the regression technique to predict drug side effects [14][15]. Uniform classification models can be utilized for prediction reactive behavior of drugs [16]–[19]. In these models, pairs of drugs are considered as samples. These pairs are considered positive if a drug has side effect, otherwise it is considered a negative sample. But among these pairs, most of the samples are negative samples i.e, doesn’t have any side effect. Selection of negative samples can affect the performance of the prediction model. In some studies, this problem can be overcome with selecting samples randomly. But this solution can be cause of neglection of several positive samples random selection.
Machine learning methods are effectively being used in medical science as well as other fields. Zhang et al. [11] proposed a model named ‘Feature Selection Based Multi-Label K-Nearest Neighbor Method’ (FS-MLKNN) that can determine simultaneously critical feature dimension and can solve the multi-label problem effectively. Further, they developed an ensemble learning model with the FS-MLKNN model to improve performance. They evaluated the proposed model on the various datasets.
Atias & Sharan [12] proposed a novel approach to keep in consideration the drugs and their side effects. The combination of Network-based diffusion and canonical correlation analysis was applied to predict the side effect of the drug. The cross-validation technique was used to evaluate the model performance. About 692 drugs were considered and the source of the dataset was Package Insert [20] in this study.
Niu & Zhang [15] proposed an effective technique named as quantitative prediction, in which they tried to predict the quantitative scores of the drugs.Especially, explored and evaluated the features of drugs for for quantitative prediction. After analysis of individual features, they considered feature combination (Chemical Structure, Targets, Treatment Indication) strategy in their study. As a final quantitative prediction model, they used the average scoring ensemble method that improved the performance for drug side effect prediction (Quantitative).
Zhao et al. [16] proposed a binary classification technique for drug side effects prediction. In this study, they took the drug and side effects as pairs and converted this problem into a binary classification. On the behalf of similarities, the pairs were represented as five features that relate to drug property. Further, they analyzed features of each drug and concluded that drug similarity in the fingerprint of the drug is the most important feature for side effect prediction.
Zhao et al. [17] adopted a network embedded method to extract informative and useful features of drugs from heterogeneous networks, that represent different properties of drugs. The features consists of requisite information of drug as well as of drug side effects. Random forest network model was used as a prediction model in this study that was able to get the average of Matthews correlation coefficients of 0.640 and 0.641 with balanced and unbalanced datasets respectively.
Ding et al. [18] proposed an effective model for drug side effect association that is based on the Multiple Learning Algorithm (MKL). MKL is built from the drug and side effect space. Finally, they used a graph-based semi-supervised technique for the construction of a side effect predictor.
Ding et al., 2019 [19] proposed an effective model for drug side effect association. They initially constructed the multiple kernels respectively from drug and side effect space that later weighted linearly in two different spaces with Centered Kernel Alignment-based Multiple Kernel Learning. To fuse the drug and side effect Kernel Kronecker Regularized Least Squares was used at the end. They evaluated their model on three benchmark datasets and were able to achieve an effective Area Under Precision-Recall Curve Score of 0.679, 0.672, and 0.675 on Mizutani’s dataset, Pauwels’s dataset, and Liu’s dataset, respectively. Summery of literature is mentioned in Table 1.
Table 1: Literature Summary
|
Author |
Model |
Method/Approach |
Description |
|
(Zhang et al., 2015) [11] |
FS-MLKNN |
Feature Selection and Ensemble Technique |
Determine the critical feature dimension and ensemble learning technique is used. |
|
(N & R, 2011) [12] |
- |
Network-Based Diffusion and Canonical Correlation Analysis |
An approach is proposed that is the combination of diffusion of network and canonical analysis in this study for side effect prediction. The 692 drugs are considered in this study and results are evaluated with cross-validation technique. |
|
(Y & W, 2017) [14] |
- |
Quantitative Prediction Approach and Ensemble Method |
The three features combination (Chemical Structure, Targets, Treatment Indication) are considered and the ensemble method is used as the final quantitative prediction model. |
|
(Zhao et al., 2018) [15] |
- |
Binary Classification Technique |
The drug and side effect consider as pairs and the problem is converted into a binary classification problem in this study. Pair is represented as five features that relate to drug properties. |
|
(Zhao et al., 2019) [17] |
- |
Network Embedded Approach |
To extract useful features (Drug properties) network embedded approach is considered, a separate network is deigning for drug side effects. And finally, features are combined for the random forest model network model. |
|
(DIng et al., 2019) [18] |
- |
Multiple Learning Algorithm and Graph-Based Semi-Supervised Technique |
An MLK algorithm that constructs with drug and side effect space is considered and a graph-based semi-supervised approach is used for the final side effect prediction. |
|
(Ding et al., 2019) [19] |
- |
CKA-MKL and Kronecker RLS |
Multiple Kernels are constructed with drug and side effect space and converted to linear weights with the optimization of the CKA-MKL algorithm in two different spaces and Kronecker RLS is used to fuse drug kernel as well side effect kernel, to identify side effect association. |
Still, there is a lack of a novel model that can predict real time side effects (e.g., fever, Cough. Dizziness, etc.). For this problem, we have proposed a deep learning-based model named “Deep Learning Model for Side Effect Prediction” (DLMSE). We have collected the side effect for drugs and labeled them as one-hot encoding to deal with our problem as a multilabel classification causing various advers reactions. The main steps involved in this study are:
- Dataset Preparation (One Hot Encoding)
- Selection of an effective method for conversion of drug chemical structure into Morgan Fingerprint (RDKit)
- DLMSE model for drug side effect prediction
- MATERIALS & METHODS
Dataset: SIDER [20] has information along with the attributed e.g., adverse reactions, side effects classifications, side effects frequency as well as information of drug-target relation. This information is extracted from public documents as well as from package inserts [19]. In this study, we have used common drugs and their side effects that have been extracted from SIDER. Initially, in this study, our focus was on three side effects that are ‘Dizziness’, ‘Allergy’ and ‘Headache’ as most of the drugs cause these side effects [20]. A drug has four features ‘Chemical Structure’, ‘Target’, ‘Enzyme’, and ‘Pathways’. The ‘Chemical Structure’ always remains consistent so in this study, we have proposed a model that can predict side effects on the behalf of ‘Chemical Structure’. We have converted the chemical structure format into ‘Morgan Fingerprint’ as a bit vector with RDKit [21] for effective interpretation.
Table 2: Parameters of RDKit for Morgan Fingerprint
|
Radius |
4 |
|
nBits |
881 |
We have scraped the data from SIDER with the python selenium library [21]. It is noticeable that a single drug can be the cause of multiple side effects, in this study we have dealt a single drug with three side effects, and one hot encoding scheme is used to set side effects in form of 0-1 vector, 0 means the absence of side effect and 1 means the presence of the side effect. We have considered 1365 drugs in this study. The encoded form of 2D/3D molecule features as in the form of the array is known as Fingerprint, simply it is a way of encoding. As the first five samples of the prepared dataset in the converted form of fingerprints are shown in Table 3.
Table 3: Prepared Dataset Samples
|
Fingerprint |
Dizziness |
Allergy |
Headache |
|
0|10|12|40|45|69|92|101|108|109|112|113|114|116|118|156|165|187|193|197|210|246|271|286|298|307|331|352|357|374|377|401|424|430|432|456|459|463|468|472|478|487|498|507|510|523|542|544|547|549|561|606|615|648|656|672|679|707|708|717|720|724|735|736|737|760|768|771|775|791|806|810|816|828|829|836|844|847|878 |
0 |
1 |
0 |
|
0|10|18|33|36|43|45|87|92|95|98|100|103|108|113|114|124|147|172|178|215|222|239|246|256|277|280|286|295|298|305|311|327|337|343|344|349|366|371|383|387|399|409|410|424|425|430|435|437|449|465|486|489|490|531|537|543|545|554|575|584|588|596|604|616|619|623|628|638|640|658|660|673|702|703|704|709|711|714|716|717|734|737|740|748|749|763|769|775|779|802|810|816|850|859|874 |
0 |
0 |
0 |
|
0|10|18|45|47|48|56|60|68|81|84|85|86|93|106|113|114|133|156|170|171|181|183|194|200|245|257|263|298|304|334|335|344|349|360|371|381|395|399|410|419|424|429|430|431|455|456|478|486|504|506|512|526|541|545|555|558|602|607|614|619|621|622|623|668|685|701|707|716|717|726|731|735|736|737|775|782|800|810|812|816|821|825|829|844|855|857 |
1 |
0 |
1 |
|
0|10|27|32|44|54|59|60|61|68|80|105|109|113|114|119|121|133|151|157|177|185|203|213|214|219|233|243|256|257|266|285|289|298|306|307|326|328|329|333|335|337|338|343|352|379|389|399|403|416|417|424|428|436|437|439|443|449|456|459|460|463|475|481|516|521|522|548|563|575|588|589|594|600|605|616|629|640|657|671|684|694|695|702|707|710|711|717|725|730|737|745|751|758|765|775|778|779|788|792|793|794|801|809|810|828|831|838|841|846|851|852|863|873|874|879 |
1 |
1 |
0 |
|
0|11|18|87|128|165|215|246|284|344|383|399|410|430|456|545|592|640|661|703|714|716|775 |
0 |
0 |
1 |
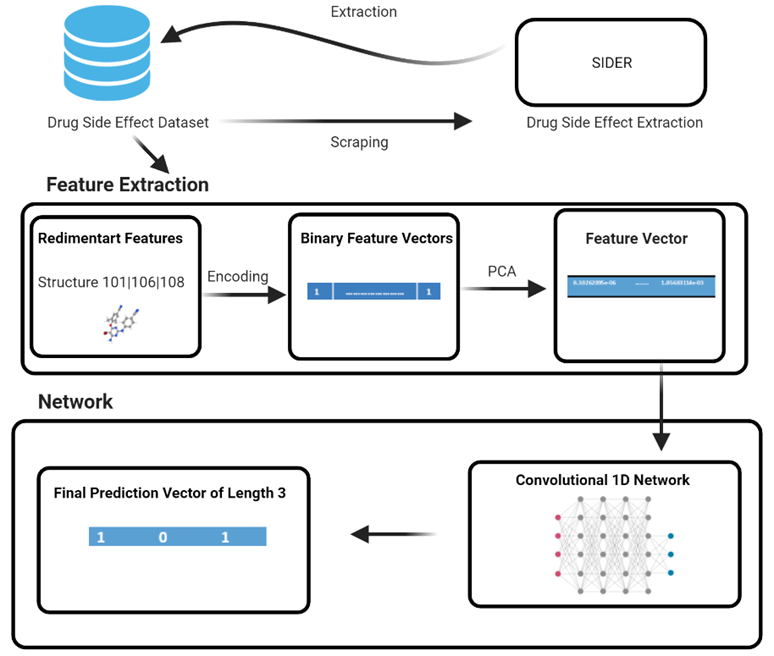 Overview of DLMSE: The purpose of this study was to propose a convolutional neural network-based model named ‘DLMSE’ for the prediction of drug side effects with drug chemical structure. Due to the 1D shape of the dataset, we have designed a Convolutional 1D neural network.
Overview of DLMSE: The purpose of this study was to propose a convolutional neural network-based model named ‘DLMSE’ for the prediction of drug side effects with drug chemical structure. Due to the 1D shape of the dataset, we have designed a Convolutional 1D neural network.
Figure 1: Pipeline of DLMSE
Figure 1 is the pipeline of the DLMSE model to achieve effective accuracy. The network is based on ‘Chemical Structure’ that is represented as a fingerprint. Firstly, the dataset is scrapped from the SIDER website. We have converted the chemical structure in integer representation with the Rdkit python library. Features are represented as a binary vector in the form of 0 and 1 and this process is called encoding. Further, we have applied Principal Component Analysis (PCA) for dimensionality reduction purposes and fed these vectors into the network for getting the output vector of length 3. A detailed description of model construction and optimization is presented below.
DLMSE Construction and Optimization:
At this stage, we have optimized features to train the model. DLMSE is based on the 1D convolutional network due to the ID shape of data. Multiple layers scheme is used in DLMSE construction. We have trained our model with the chemical structure to predict the side effects of drugs in the future. The activation function is also known as the transfer function used to get the output of the node. Currently the most useable activation function is Rectified Linear Unit (ReLU) [23]. It can be used in all convolutional neural networks as well as for deep learning, we have also considered it as an activation function in our proposed model. As it can be seen in Equation 1 R(z) will be 0 if z is less than 0 otherwise it is equal to z.
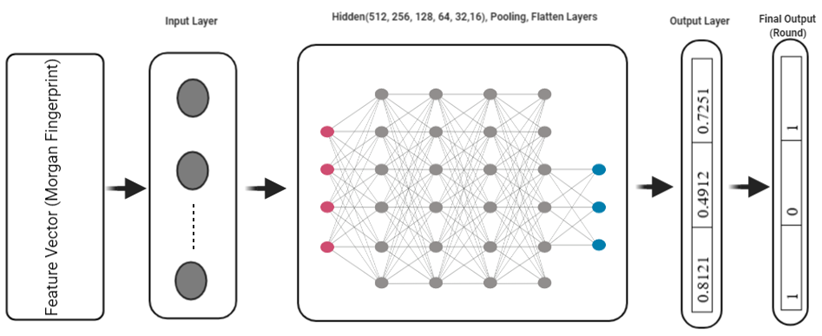
Figure 2: DLMSE Model Construction
Feature Selection:
In machine learning, more training samples can lead to an effective machine learning model but this statement is not correct in terms of features. The data that we collected from the real world has a lot of meaningless features that put a negative impact on the performance of the model. It is necessary to select necessary features and ignore the unnecessary features to build an effective model in terms of accuracy as well as computation time. In this study, we have used PCA that is a technique of dimensionality reduction. Matrix factorization plays an important role in PCA to reduce the dimensionality of the dataset. It can be used to reduce the features and is best suitable where the dimension of the dataset is high. We can set the required number of features as ‘n_components’ as for our problem we have set it to 500 to get 500 useful features from the total of 881 features after the experiments.
Evaluation Metrics:
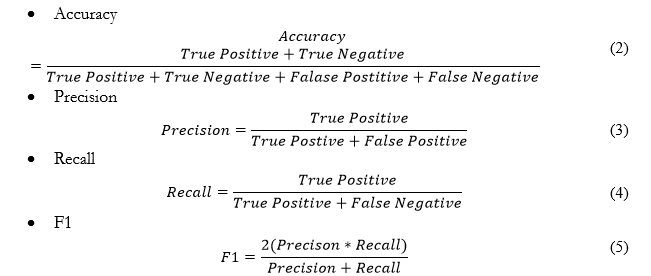
As in the medical field, it is difficult to collect too much data especially in the low economy country, and on some samples, we cannot say that our model is good and effective for the real world. To overcome this problem, we have used the “RepeatKFold” method to train and evaluate the model with different folds at each iteration. We have set ‘n_splits=10’, ‘n_repeat=5’ and ‘random_state=42’ as arguments for RepeatKFold. Moreover, due to multiple side effects of a single drug, we deal with our problem as a multi-label problem. We have considered Accuracy, Precision, Recall, F1, and AUC scores as evaluation scores.
Parameters Optimization:
In neural networks, parameter optimization is compulsory to reduce the computational cost of the model as well as to reduce the training time of the model. We need to tune the dropout rate, learning rate, number of epochs, batch size, learning rate, and hidden layers. We have considered the different number of hidden layers in this study and finally conclude that 6 hidden layers 512, 256, 128, 64, 32, 16 respectively are effective in this study. Further, we have also considered different dropout rates as well as the learning rate and conclude that dropout of 0.001 and learning rate of 0.3 give us effective results as compared to other values that can be seen in Table 4.
Table 4: Accuracy of the proposed model with different parameters
|
Feature |
Learning Rate |
Dropout Rate |
Accuracy |
|
Chemical Substructure |
0.1 |
0.01 |
0.8339 |
|
Chemical Substructure |
0.001 |
0.1 |
0.9422 |
|
Chemical Substructure |
0.01 |
0.001 |
0.8474 |
|
Chemical Substructure |
0.001 |
0.2 |
0.9452 |
|
Chemical Substructure |
0.001 |
0.3 |
0.9494 |
Results
We have achieved an accuracy of 0.9494 and an AUC score of 0.9698 with our proposed model for drug side effects prediction with drug chemical structure. The model is validated with different train test split with the RepeatKFold method. All the other considered evaluation scores are effective of our proposed model. The proposed model results are shown in Figure 3.
 Figure 3: Mean Evaluation Scores of Proposed Model
Figure 3: Mean Evaluation Scores of Proposed Model
For the robustness of our model, we have compared it with other machine learning models e.g., Logistic Regression (multi_class='multinomial', solver='lbfgs'), SVM, Random Forest, KNN(k=3) with “MultiOutputClassifier”. Table 5 shows the robustness of our model in terms of evaluation scores comparison with machine learning models. Table 5 shows that the performance of our proposed model is effective than other machine learning models.
Table 5: Robustness of Proposed Model
|
Model |
Accuracy |
AUC |
Precision |
Recall |
F1 |
|
DLMSE |
0.9494 |
0.9698 |
0.9850 |
0.9850 |
0.9840 |
|
Logistic Regression |
0.4775 |
0.5584 |
0.7779 |
0.9237 |
0.8437 |
|
SVM |
0.4287 |
0.5738 |
0.7871 |
0.8629 |
0.8224 |
|
Random Forest |
0.5128 |
0.5222 |
0.7612 |
0.9839 |
0.8569 |
|
KNN |
0.4402 |
0.5440 |
0.7717 |
0.9081 |
0.8336 |
After applying the model as well as other well-known machine learning model on a prepared dataset, we have come to the point that our proposed model is effective than other machine learning models. This model is effective in terms of considered evaluation metrics as can be seen in Table 5.
Most of the studies exist that are about the DDI (Drug-Drug Interaction). DDI is a reaction (side effects) that occur when two or more drugs are taken at the same time and these side effects are very dangerous for human lives. But few studies [24][25][26] focus on real side effects like fever, flu, headache, dizziness that we faced in our daily life. There should be a model that can diagnose real-life side effects. So, we have proposed a convolutional 1D base model as our problem is one-dimensional. RDKit python library is considered in this study to convert Chemical Substructure to Morgan Fingerprint.
Discussion
In this study, we have considered 3 side effects (Dizziness, Headache, Allergy) in terms of multi-label classification as 1 drug can cause multiple side effects. We use medicine on regular basis but most of the time these medicines cause side effects and due to major side effects, we lose the lives of our beloved ones. This study is useful for the pharmaceutical companies to launch a drug that has no major side effects because if the drug has major side effects, then immediately it needs to be withdrawn from the market. Thus,it can cause a huge loss to pharmaceutical companies in terms of time and money.
To keep the importance of this study in terms of human lives and pharmaceutical companies, we have proposed a deep learning-based model that can predict the side effects of drugs with chemical substructure (Smile – Alternative name of chemical substructure). We have prepared the dataset with the help of the SIDER website; we have scrapped the dataset with side effects as a key of search. We have performed the experiments with different hyperparameters as illustrated in Table 4.
Recently, deep learning is used in the drug discovery field, we have applied PCA (Principal Component Analysis) with the proposed model to select effective features and to optimize the complexity of the proposed model as well as to avoid the overfitting problem. We have also considered the well know hyperparameters e.g., Learning Rate and Dropout Rate that needs to be optimized in designing a deep learning-based model.
The limitations of this study are the smaller number of samples. To overcome the problem, we have considered different Repeat KFold methods to train and evaluate the model on the different split.
Conclusion
This study proposed an effective model for the prediction of drug side effects with drug chemical substructure. Based on the concept that a single drug can cause multiple side effects; we deal with our problem as a multilabel classification problem. This PCA technique is used for feature dimensionality reduction as well as dropout to avoid overfitting. This model can effectively predict three side effects (‘Dizziness’, ‘Allergy’ and ‘Headache’) of drugs. At the same time, this model can predict the side effects of a candidate drug to reduce the cost and time of the drug discovery process. Compared with state-of-the-art methods of machine learnings (Logistic Regression, Support Vector Machine, Random Forest, K-Nearest Neighbor), our proposed model (DLMSE) produces more effective results for the three side effects dataset.
ACKNOWLEDGEMENT
Author’s Contribution. All the authors contributed equally.
Conflict of interest. We declare no conflict of interest for publishing this manuscript in IJIST.
Project details. Nil
Dataset & Codes: Provided on Request.
REFERENCES
[1] “Rofecoxib,” Meyler’s Side Eff. Drugs, pp. 236–239, 2016, doi: 10.1016/B978-0-444-53717-1.01416-5.
[23] J. Bjorck, C. Gomes, B. Selman, and K. Q. Weinberger, “Understanding Batch Normalization.”




















