Evaluation of Microbial Contamination in Meat and its Control Using Preservatives
Rabia Rehman1, Sumaira Mazhar1*, Mawra Gohar1
1 Department of Biology, Lahore Garrison University
* Correspondence Author’s Email ID: Dr. Sumaira Mazhar, smz.mmg@gmail.com
Citation | Rehman. R, Mazhar. S, Gohar. M, “Evaluation of Microbial Contamination in Meat and Its Control Using Preservative”. International Journal of Innovations in Science and Technology. Vol 4, Issue 2, 2022, pp: 404-415.
Received | March 31, 2022; Revised | April 24, 2022, Accepted | April 25, 2022; Published | April 27, 2022.
________________________________________________________________________
Food-borne illnesses are common in both developed as well as developing countries. The majority of foodborne diseases are caused by consuming contaminated meat products. This study aimed to evaluate the microbial contamination in different meat samples. Chicken (n=20), Mutton (n=20) and beef (n=20) samples were collected from 10 towns. Total viable count (TVC) and Total coliform count (TCC) in different meat samples were checked. Microscopic, macroscopic and biochemical profiling of isolates (n=108) was done. It was observed that E. coli was the more common (34%) pathogenic bacteria found in raw chicken followed by Salmonella (28%), Staphylococcus (25%), Shigella (8%), Enterobacter (2%), and Bacillus (3%). In Beef Samples E. coli (39%) was more common followed by Salmonella (30%), Staphylococcus (18%) and Enterobacter (8%), and Shigella (5%). While in Mutton Samples E. coli (32%), Salmonella (32%), Staphylococcus (12%), Shigella (12%), Enterobacter (9%), and Bacillus (3%). Antibacterial activity of natural preservatives i.e., Ginger, Garlic, and Radish, and commonly used synthetic preservatives i.e., Sodium nitrite was also checked on isolated strains. It was observed that Ginger and Garlic showed maximum antibacterial activity at the highest concentration used up to 0.8g/ml. Radish showed no antibacterial activity at any concentration. Antibacterial activity of Sodium nitrite was also higher at the maximum concentration used (0.006mM). The renowned harmful effects of Sodium nitrite, make it necessary to devise the use of natural preservatives. It was observed that ginger and garlic may serve as natural preservatives for meat preservation without any side-effect. However, more research is required for the implementation of natural preservatives for meat storage and safety.
Keywords: Microbial load, raw meat, natural preservatives, antibacterial activity, meat spoilage.
INTRODUCTION
Meat is a rich source of protein, fatty acids, minerals, and various vitamins [1]. It contains all the nine essential amino acids required for optimal human health [2]. It is the most commonly used food in developed as well as developing countries. It contains certain components (fats, proteins, and carbohydrates) that make it more prone to catch microorganisms that are potent pathogens. The meat itself is free of contamination when obtained from healthy animals. However, contamination is more likely to occur during slaughtering, handling, cutting, and transportation. Contaminated meat poses a huge risk to human health. Microbial contamination is the most hazardous type of contamination that is known for causing food-borne diseases.
Food-borne illnesses are more prevalent in developing countries including Pakistan [3]. Meat-borne prionic, viral, bacterial, protozoal, parasitic, and fungal diseases are well known [4]. Heterogenous microbial flora present in red and white meat is the main cause of diseases including zoonotic diseases. Pathogen present in meat causes gastrointestinal infections including diarrhea and hemolytic uremic syndrome [1]. Foodborne pathogens are a major threat worldwide. Different conducted studies showed the increasing trend of harmful foodborne contaminants in a variety of local foods. [5]. A total of 800 most common food samples such as chicken, beef, raw milk, vegetables, and salad samples were collected in the retail market, the total contamination of food samples and infected workers was 48.37%. Food samples were also found to be suitable for Salmonella spp. (19%), E. coli (O157: H7) (8%) and 1.25% Listeria monocytogenes. 5% of tested food samples were found to be infected with at least two viruses. The results urge the adoption of appropriate food hygiene practices to reduce the incidence of foodborne disorders [6].
Meat is preserved by hindering microbial growth, avoiding fatty acid oxidation, and abating the enzymatic activity. Different techniques are used to preserve meat by avoiding meat spoilage, including chilling, freezing, meat curing, meat smoking, canning, drying, irradiation, fermentation, and vacuum packaging [4]. Some synthetic preservatives are also used for meat preservation. These preservatives include sodium nitrite, potassium nitrite, nisin, potassium sorbate, etc. [7]. Synthetic preservatives are well known for their antimicrobial activity as they minimize lipid oxidation by reinstating free radicals [8]. Despite many advantages, synthetic preservatives are not appreciated by consumers due to their potentially harmful effects. Nitrites and nitrates have the potential to cause bladder cancer, colon cancer, and leukemia [9]. Synthetic antioxidants such as butylated hydroxyanisole (BHA) and butylated hydroxyl toluene (BHT) has strong toxicity and carcinogenic effect [10].
Natural plant extracts are being considered a substitute for synthetic preservatives in raw meat [11]. Plants are rich in flavonoids, polyphenols, and phenolics which have antibacterial activity against many Gram-positive and Gram-negative bacteria. The antibacterial activity of rosemary, sage, thyme, oregano, chestnut, grapefruit seed, cinnamon, and turmeric is already well known [9]. Natural antioxidants found in plants and fruits get a major boost in their role in preventing auto-oxidation in fatty foods. The rhythmic leaves (Moringa oleiofera) are a powerful source of phenolics and have a high neutraceutical value to increase the performance of meat products that are very commercially important [12].
Meat is the most preferable and common food consumed in Pakistan. The poor sanitation conditions of slaughterhouses and butcher shops increase the risk of contamination in raw meat. Many studies have been conducted to evaluate microbial load in meat samples taken from specific areas of Lahore, Pakistan. However, no extensive study has been done to evaluate microbial contamination in meat from all the districts of Lahore, Pakistan. Therefore, in the presented work, we aimed to evaluate the microbial load in raw meat samples collected from different areas of Lahore. Natural preservatives are considered to be a good option as they have antibacterial activity without any side effects on human health. So, we also accessed the antibacterial activity of selected natural preservatives including Ginger, Garlic, and Radish.
MATERIAL AND METHODS
Sample collection: Meat samples (n=60) were collected from different butcher shops in Lahore. Chicken (n=20), mutton (n=20), and beef (n=20) samples were collected from different butcher shops in 10 towns of Lahore including Aziz Bhatti town, Data Gunj Bakhsh Town, Gulberg town, Iqbal town, Nishter town, Samanabad town, Shalimar town, Ravi town, Wahga town and Lahore Cantonment. Each sample was washed and cut into fine pieces (10g). Samples were labeled as CS1, CS2......CS20, BS1......BS20 and MS1...MS20 for chicken, beef, and mutton, respectively according to the towns from which these were collected. 1g of each meat sample was homogenized using a vortex machine in 10mL distilled water. After homogenization, serial dilutions of samples were prepared.
Enumeration, isolation, and identification of microorganisms
The total viable bacterial count was performed using the spread plate technique on nutrient agar. A total coliform test was performed using the spread plate method on MacConkey agar. CFU/ml was calculated using the following formula:
CFU/ml = (no. of colonies x dilution factor) / volume of culture plate
Each isolated bacterial colony was purified by inoculating it onto nutrient agar. The results were observed and noted for further analysis. The isolated bacteria were subjected to identification using standard microbiological techniques including Gram’s staining, Spore staining, etc. The isolates were then subjected to differential and selective media according to a scheme by Bergey’s manual [13]. Gram positive bacilli were streaked on blood agar plates to determine the hemolytic ability of bacterial isolates. Gram-positive cocci were streaked on Mannitol salt agar (MSA) plates to differentiate between mannitol fermenters and non-fermenter bacteria. Gram-negative bacterial isolates were streaked on MacConkey agar to check the ability of bacteria to ferment lactose and to differentiate between lactose fermenters and non-fermenters. The bacterial isolates from MacConkey agar were streaked on Eosin methylene blue (EMB) agar for further identification. The off-white bacterial colonies were further inoculated on Salmonella Shigella (SS) agar. The results were observed and noted after incubation [13]. Biochemical tests including catalase test, oxidase test, indoles test, methyl red test, Voges Proskauer test, and simmon citrate tests were also performed to identify the isolated strains.
Antimicrobial activity of natural and synthetic preservatives against bacterial isolates
Selected preservatives were washed with tap water. Ginger, garlic, and radish were peeled off, and paste was prepared using a pestle and mortar.0.2g/ml, 0.4g/ml, 0.6g/ml, and 0.8g/ml dilution of selected preservatives were made and their antibacterial activity was accessed. A commonly used synthetic preservative (Sodium nitrite) was also selected to check antibacterial activity against different isolates. Different concentration of sodium nitrite (0.2mM, 0.4mM, 0.6mM and 0.8mM) were prepared and antibacterial activity was checked. The antimicrobial activity was assessed by the disc diffusion technique [14].
RESULTS
Total viable counts (TVC) and Total coliform count (TCC)
TVC was done using the spread plate method and resultant colonies were counted using a colony counter. It was observed that chicken samples collected from Iqbal town, Shalimar town, and Lahore cantonment have more CFU/ml as compared to other towns of Lahore i.e., (2.7 x109CFU/mL). While the chicken sample taken from Samanabad town had the lowest CFU/ml (4.8 x106). Beef samples from Waghatown showed more bacterial load (2.7 x 109CFU/ml) while a low bacterial count was observed in samples collected from Shalimar town (2.9 x 106 CFU/mL). TVC estimate observed in mutton samples from Wagha town was 2.5 x 109CFU/mL while less CFU/ml was observed in mutton samples taken from Ravi town (1.3 x 107).
It was observed that chicken samples taken from Nishter town were more contaminated with total coliform (9.8 x 108 CFU/ml) than other samples collected from different towns e.g., Data Gunj bakhsh town 5.4 x 106 CFU/ml. Beef samples from Ravi town showed more CFU/ml (6.6 x 108) than other towns e.g., Lahore cantonment contained less CFU/ml (5.1 x 106). While TCC was higher in mutton samples collected from Lahore cantonment (4.5 x 108). It was observed that the mutton sample taken from Nishter town had less CFU/ml (5.3 x 106) than other samples from different towns. Overall, it was observed that the chicken sample was more contaminated as compared to mutton and beef samples as indicated in Table 1.
Table 1. Total viable counts (TVC) and Total coliform counts (TCC) of meat samples (chicken, beef and mutton)
|
Chicken |
Beef |
Mutton |
||||||
|
Sample ID |
TVC (CFU/ml) |
TCC (CFU/ml) |
Sample ID |
TVC (CFU/ml) |
TCC (CFU/ml) |
Sample ID |
TVC (CFU/ml) |
TCC (CFU/ml) |
|
CS1 |
6.4 x 106 |
6.9 x 106 |
BS1 |
1.8 x 107 |
1.3 x 107 |
MS1 |
2.2 x 109 |
1.3 x 107 |
|
CS2 |
4.7 x 108 |
3.5 x 108 |
BS2 |
1.3 x 107 |
9.4 x 106 |
MS2 |
2.8 x 107 |
1.6 x 107 |
|
CS3 |
2.2 x 107 |
5.4 x 106 |
BS3 |
2.5 x 107 |
5.4 x 106 |
MS3 |
1.9 x 109 |
1.2 x 107 |
|
CS4 |
2.2 x 107 |
8.5 x 106 |
BS4 |
1.5 x 109 |
1.6 x 107 |
MS4 |
2.9 x 107 |
1.4 x 107 |
|
CS5 |
2.5 x 107 |
1.1 x 107 |
BS5 |
2.9 x 106 |
8.2 x 106 |
MS5 |
1.2 x 109 |
1.1 x 107 |
|
CS6 |
5.4 x 108 |
5.2 x 107 |
BS6 |
2.1 x 109 |
1.2 x 107 |
MS6 |
2.3 x 109 |
9.6 x 106 |
|
CS7 |
1.0 x 107 |
1.3 x 107 |
BS7 |
2.3 x 107 |
5.2 x 106 |
MS7 |
2.7 x 107 |
1.3 x 107 |
|
CS8 |
2.7 x 109 |
6.0 x 108 |
BS8 |
1.1 x 107 |
6.4 x 106 |
MS8 |
1.9 x 109 |
1.1 x 107 |
|
CS9 |
6.9 x 106 |
9.4 x 108 |
BS9 |
1.7 x 107 |
7.1 x106 |
MS9 |
1.5 x 107 |
5.3 x 106 |
|
CS10 |
1.7 x 107 |
2.2 x 107 |
BS10 |
2.3 x 107 |
9.2 x 106 |
MS10 |
1.8 x 109 |
1.2 x 107 |
|
CS11 |
4.8 x 106 |
4.5 x 107 |
BS11 |
1.2 x 107 |
4.2 x 108 |
MS11 |
9.4 x 108 |
1.4 x 107 |
|
CS12 |
1.1 x 107 |
4.5 x 107 |
BS12 |
1.9 x 107 |
1.3 x 107 |
MS12 |
2.3 x 107 |
1.2 x 107 |
|
CS13 |
2.2 x 107 |
1.4 x 107 |
BS13 |
1.9 x 109 |
1.8 x 107 |
MS13 |
1.8 x 109 |
1.5 x 107 |
|
CS14 |
1.4 x 109 |
1.3 x 107 |
BS14 |
2.3 x 107 |
1.4 x 107 |
MS14 |
2.2 x 109 |
1.4 x 107 |
|
CS15 |
1.6 x 107 |
6.9 x 106 |
BS15 |
2.2 x 107 |
7.0 x 106 |
MS15 |
1.3 x 107 |
6.5 x 106 |
|
CS16 |
6.2 x 106 |
3.2 x 108 |
BS16 |
7.8 x 106 |
6.6 x 108 |
MS16 |
1.6 x 107 |
5.6 x 106 |
|
CS17 |
8.4 x 108 |
1.3 x 107 |
BS17 |
2.8 x 107 |
1.7 x 107 |
MS17 |
1.9 x 107 |
7.6 x 106 |
|
CS18 |
2.3 x 107 |
7.6 x 106 |
BS18 |
2.7 x 109 |
1.5 x 107 |
MS18 |
2.5 x 109 |
1.7 x 107 |
|
CS19 |
2.8 x 107 |
9.4 x 106 |
BS19 |
1.2 x 109 |
1.1 x 107 |
MS19 |
1.1 x 109 |
1.2 x 107 |
|
CS20 |
2.7 x 109 |
1.3 x 107 |
BS20 |
1.8 x 109 |
5.1 x 106 |
MS20 |
1.6 x 109 |
4.5 x 108 |
Isolation and purification of bacterial isolates from different meat samples
Each collected sample was inoculated in broth for initial enrichment. After incubation, all test tubes (100%) showed turbidity. The isolated bacteria were further inoculated in nutrient agar. After incubation, different bacterial colonies were observed. The resultant growth of nutrient agar was subjected to the purification of isolates. A total of n=108 isolates were purified from a different sample (n=60). Out of the total 108 bacterial isolates, 36 (33.3 %) isolates were purified from chicken samples, 37(34.2%) isolates were purified from beef while 35 (32.4%) were isolated from mutton samples.
Microscopic characterization of bacterial isolates
Gram staining of each bacterial isolate was performed. It was observed that 35 (32%) isolates were Gram-positive while 73 (68%) were Gram-negative. Among 108 strains, 81(75%) were rods while 27 (25%) were cocci arranged in diplococci, chains or clusters. In the chicken sample, Gram-positive Cocci 9 (25%), Gram-positive rod 1(2.7%), Gram-negative rod 26(72.2%) were observed while in the mutton sample, Gram-positive Cocci 8(22.8%), Gram-positive rod 1(2.8%), Gram-negative rod 26 (74.25%) were observed. Beef samples showed Gram-Positive Cocci 10(27%), Gram-positive rod 0(0%), and Gram-negative rod 27 (72.9%) bacterial isolates. Spore staining was also performed to differentiate spores between former Gram-positive bacteria and non-spore former. Out of 2 Gram-positive rods, a Total of 2 (100%) bacterial isolates were spore former.
Macroscopic characterization of bacterial isolates
Gram-negative (rods and cocci) bacterial isolates (n=73) were streaked on MacConkey agar to differentiate between lactose fermenters and lactose non-fermenters. It was observed that out of 26 Gram-negative bacteria isolated from chicken, 13 (50%) showed pink colonies on MacConkey agar while the other 13 (50%) were off-white. In beef samples, 14 (52%) showed pink colonies on MacConkey agar while 13 (48%) showed off-white growth on MacConkey agar. From mutton samples, the pink and off-white colonies showed lactose fermentation and no fermentation in 43.3% and 57.69% respectively. It was observed that in beef samples, more bacterial isolates (52%) showed pink colonies on MacConkey agar while bacteria isolated from mutton samples (57.69%) were observed as non-lactose fermenters on MacConkey agar.
Pink colonies grown on MacConkey agar were further streaked onto Eosin methylene blue (EMB) agar for further confirmation. It was observed that among 38 strains showing pink colonies, 34 (90%) bacterial isolates showed green metallic sheen on EMB agar while 4 (10%) isolates showed no growth on EMB agar. Out of 13 bacteria isolated from chicken samples, 11 strains (85%) showed metallic green color on EMB agar while 2(18%) isolated bacteria showed no growth on EMB agar. All the isolated bacteria (100%) taken from beef samples were observed in the form of a green metallic sheen on EMB agar. Total 9 bacterial isolates (82%) picked from mutton samples showed metallic green color on EMB agar.
Gram-positive cocci were streaked on mannitol salt agar to check the ability of bacteria to ferment mannitol. It was observed that out of 9 Gram-positive cocci isolated from chicken, 9(100%) bacterial isolates showed yellow growth. Among 10 bacteria isolated from beef, 7 (70%) showed positive growth on MSA. 4 strains (50%) isolated from mutton indicated positive growth on MSA which indicated that these bacterial isolates are mannitol fermenters and produce yellow colonies.
Bacterial isolates were streaked on blood agar and it was observed that about 72% of bacterial isolates showed beta hemolysis while 28% of bacterial isolates showed gamma hemolysis on blood agar. It was observed that among Gram-positive bacteria isolated from chicken, 100% isolate showed beta hemolysis. 70% of bacterial isolates from beef samples showed beta hemolysis while 30% of bacterial isolates showed gamma hemolysis. 50% of gram-positive bacterial isolates from mutton samples indicated beta hemolysis while 50% of bacterial isolates showed gamma hemolysis.
Off-white colonies from MacConkey agar were streaked on Salmonella Shigella (SS) agar for further identification of bacterial isolates. Plates were incubated for 24 hours at 37⁰ C. It was observed that among 13 bacterial isolates (off-white colonies on MacConkey) from chicken samples, 10 (78%) bacterial isolates showed black centered colonies on SS agar while 3 (22%) were colorless on the same agar. Among 13 selected bacterial isolates from beef, 11(85%) bacterial isolates showed black-centered colonies on SS agar while 2 (15%) showed colorless colonies. Out of 15 bacteria isolated from mutton samples, 11 (73%) isolates were with black colonies while 4 (27%) appeared as colorless colonies on SS agar. It was observed that in chicken samples, more bacterial isolates (78%) showed black colonies on SS agar as compared to beef and mutton while in mutton samples, more bacterial isolates (22%) showed colorless colonies on SS agar.
Biochemical profiling of isolates
Biochemical tests were performed to confirm the identification of selected organisms. Citrate, oxidase and Indole, the Methyl Red, the Voges-Proskauer and the Citrate Test (IMViC) tests were performed. A catalase test was performed on all isolates while an oxidase test was performed on gram-negative bacterial isolates. IMViC tests were also performed to differentiate members of coliform.
Among 36 isolated bacteria from chicken samples, all (100%) were catalase-positive, 9 bacterial isolates (25%) were coagulase-positive and 36 isolates (100%) were oxidase negative. 10 bacterial isolates (27%) showed a positive citrate test while 73% gave negative results on citrate agar. 16 isolated bacteria (44%) showed positive indole test while 20 isolated bacteria (56%) indicated negative indole test. 97% of bacterial isolates showed positive methyl red test while 10 bacterial isolates (27%) showed positive Voges-Proskauer (VP) results.
It was observed that among 37 bacterial isolates from beef samples, 34 isolates (92%) were catalase-positive, 7 bacterial isolates (18.9%) were coagulase-positive while all isolated bacteria (100%) were oxidase negative. 7 isolated bacteria (19%) showed a positive citrate test. 16 bacterial isolates (43%) showed positive indole test, 34 isolated bacteria (92%) showed positive Methyl red (MR) results and 10 bacterial isolates (27%) were VP positive.
Biochemical profiling of isolated bacteria from mutton samples indicated that among 34 strains, 31 strains (91%) were catalase-positive, 4 strains (11.7%) were coagulase-positive and all strains were oxidase negative. 7 strains (21%) were citrate positive, 14 strains (41%) were indole positive, 30 strains (88%) were MR positive and 8 strains (23%) were VP positive.
Microbial identification of isolated bacteria
Macroscopic, microscopic characterization, and biochemical tests were performed to confirm the identification of selected organisms. It was observed that E. coli (34%) was the more common pathogenic bacteria found in raw chicken followed by Salmonella (28%), Staphylococcus (25%), Shigella (8%), Enterobacter (2%), and Bacillus (3%). In beef samples, E. coli (39%) was more common followed by Salmonella (30%), Staphylococcus (18%) and Enterobacter (8%), and Shigella (5%). While in mutton samples E. coli (32%), Salmonella (32%), Staphylococcus (12%), Shigella (12%), Enterobacter (9%), and Bacillus (3%) as indicated in Table 2.
Table 2. Total percentage of different bacterial isolates from meat sample (beef, mutton, and chicken)
|
Name of bacteria |
Beef (%) |
Mutton (%) |
Chicken (%) |
|
E. coli |
39 |
32 |
34 |
|
Staphylococcus |
18 |
12 |
25 |
|
Salmonella |
30 |
32 |
28 |
|
Shigella |
5 |
12 |
8 |
|
Enterobacter |
8 |
9 |
2 |
|
Bacillus |
0 |
3 |
3 |
Antibacterial activity of natural and synthetic preservatives
Antibacterial activity of natural preservatives (ginger, garlic, and radish) and the synthetic preservative (sodium nitrite) was observed. Strains were selected based on their abundance in meat samples. E. coli was picked from a sample BS20 plate. Staphylococcus, Salmonella, Shigella, Bacillus, and Enterobacter strains were selected from CS20, MS3, MS5, MS9, and MS4 respectively. It was observed that E. coli was more prevalent in beef samples, in chicken samples Staphylococcus was more prevalent while Salmonella, Bacillus, Shigella, and Enterobacter were more prevalent in mutton.
Antibacterial activity of ginger against different isolates
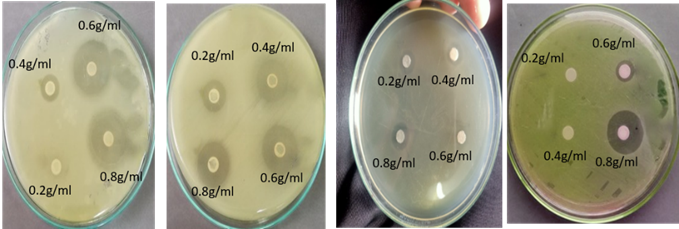
Antimicrobial activity of ginger was assessed against different isolates at different concentrations (0.2g/ml, 0.4g/ml, 0.6g/ml and 0.8g/ml). 24 hours fresh cultures (in Nutrient Broth NB) of both strains were used. It was observed that ginger shows maximum antibacterial activity against E. coli, Staphylococcus, Salmonella, Shigella, Enterobacter, and Bacillus at 0.8g/ml concentration. Figure 1 shows the size of inhibition zones against different isolates.

Figure 1. The Antimicrobial effect of ginger against isolated pathogens
|
a |
b |
c |
d |
Figure 2. Antibacterial activity of ginger a) Effect of ginger extract on (b) Effect of ginger extract on Staphylococcus, (c) Effect of ginger extract on Salmonella, (d) Effect of ginger extract on Enterobacter
Antibacterial activity of garlic against different isolates
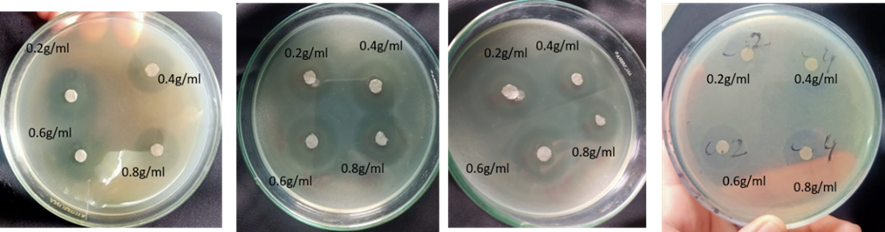
Antimicrobial activity of garlic was also assessed different isolates at different concentrations (0.2g/ml, 0.4g/ml, 0.6g/ml and 0.8g/ml). It was observed that garlic shows maximum antibacterial activity against E. coli, Staphylococcus, Salmonella, Shigella, Enterobacter, and Bacillus at 0.8g/ml concentration. Figure 3 shows different inhibition zones against different isolates.
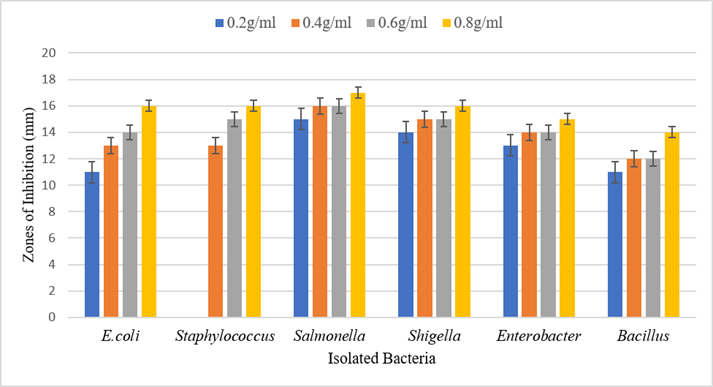
Figure 3. The antimicrobial effect of garlic against isolated pathogens

|
a |
b |
c |
d |
Figure 4: Antibacterial activity of garlic (a) Effect of Garlic extract on E. coli, (b) Effect of Garlic Extract on Staphylococcus, (c) Effect of Garlic extract on Salmonella, (e) and Effect of Garlic extract on Enterobacter.
Antibacterial activity of radish against different isolates
Antimicrobial activity of radish was also assessed on different isolates at different concentrations (0.2g/ml, 0.4g/ml, 0.6g/ml and 0.8g/ml). It was observed that radish show no antibacterial activity against E. coli, Bacillus, Staphylococcus, Salmonella, Shigella, and Enterobacter at any concentration.
Antibacterial activity of sodium nitrite against different isolates
The antimicrobial activity of synthetic preservatives (sodium nitrite) was assessed against different isolates. Antibacterial activity was checked at different concentration of sodium nitrite (0.2mM, 0.4mM 0.6mM and 0.8mM). It was observed that sodium nitrite shows maximum antibacterial activity against E. coli, Staphylococcus, Salmonella, Shigella, Enterobacter, and Bacillus at 0.8mM concentration. Figure 5. shows the size of inhibition zones against different isolates.
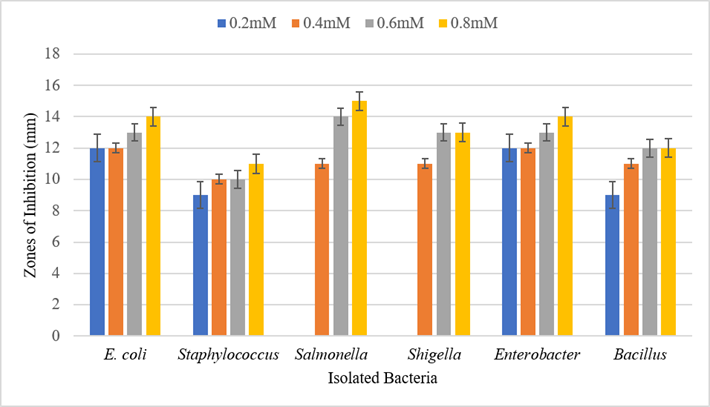
Figure 5. Antimicrobial effect of sodium nitrite against isolated pathogens
|
a |
b |
c |
d |
Figure 6. Antibacterial activity of Sodium nitrite (a) Effect of Sodium Nitrate on E. coli, (b) Effect of Sodium Nitrate on Staphylococcus, (c) Effect of Sodium Nitrate on Salmonella, (d) Effect of Sodium Nitrate on Shigella
DISCUSSION
Food-borne diseases are very common in developing countries with high prevalence and mortality rates. Among foodborne diseases, a large proportion is caused by contaminated meat or meat products [15]. As meat is obtained from poultry, fish, cattle, etc. probability of zoonotic infections is high. Raw meat is considered to be more contaminated as compared to ready-to-eat meat as reported by Kumari and co-researchers in 2019 [16]. The spoilage and low quality of meat are associated with different physical and biological factors. Heat and moisture are common physical factors that play a significant role in food spoilage. Along with the physical parameters, the prevalence of microorganisms plays a major role in meat spoilage and lowering the quality, these microorganisms include coliforms, fecal coliforms, and S. aureus, Salmonella, Shigella, listeria, etc. [17]. Unhygienic conditions and poor storage conditions are two main causes of food-borne illnesses. Meat-borne diseases are classified into different groups like prionic disease, protozoal disease, fungal disease, parasitic disease, and most common meat-borne bacterial diseases.
Therefore, it was aimed to detect bioburden in different meat samples. The presented research aimed to access the antibacterial activity of some natural preservatives (ginger, garlic, and radish) and one commonly used synthetic preservative (sodium nitrite). Different meat samples (n=60) i.e., chicken, beef, and mutton (n =20 from each type) were collected from different towns of Lahore city including Aziz Bhatti town, Data Gunj Bakhsh Town, Gulberg town, Iqbal town, Nishter town, Samanabad town, Shalimar town, Ravi town, Wahga town and Lahore Cantonment. These samples were processed and different dilutions were made to check the TVC and TCC in different meat samples. Nutrient agar plates were used to check colony morphology. Different colonies were picked for gram staining and a specific colony identity (ID) was assigned to all isolates. Spore staining was done on all Gram-positive rods to check spore formers. Gram-negative isolates were incubated at MacConkey agar and pink colonies from MacConkey agar were streaked at EMB agar while off-white colonies were streaked at SS agar. Gram-positive cocci were incubated at MSA. Blood agar was used to identify the alpha, beta, and gamma hemolytic bacteria. Isolates were identified and confirmed by biochemical profiling.
Antibacterial activity of natural and synthetic preservatives was checked on isolated strains using their different concentrations by the disc diffusion method. TVC of all samples was calculated. It was observed that mutton samples were more contaminated as compared to chicken and beef samples. A study conducted in Karachi, Pakistan indicated that beef samples were more contaminated followed by chicken and mutton [18]. TCC of all the samples was counted. It was observed that TCC was higher in chicken samples as compared to mutton and beef samples. Another study conducted in Karachi also reported that almost all the samples (chicken, beef, and mutton) are contaminated with coliform which are indicators of poor sanitation [18].
A total of n=108 isolates were purified from a different sample (n=60). Out of the total 108 bacterial isolates, 36 (33.3 %) isolates were purified from chicken samples, 37(34.2%) isolates were purified from beef while 35 (32.4%) were isolated from mutton samples. Gram staining of isolates showed that Gram-negative bacteria were more prevalent than Gram-positive bacteria in all meat samples. Our results were in concordance with research conducted in Hyderabad which reported a relatively high prevalence of Gram-negative bacteria in different meat samples (chicken, mutton, beef, fish, etc.) [19].
Gram-negative (rods and cocci) bacterial isolates (n=73) were streaked on MacConkey agar to differentiate between lactose fermenters and lactose non-fermenters. It was observed that among isolates from chicken samples, lactose fermenters and non-lactose fermenters were equal. In beef samples, more bacterial isolates (52%) were lactose fermenters while in mutton samples, non-lactose fermenters were more prevalent (57.69%). 38 strains showing pink colonies were incubated on EMB agar and 34 (90%) bacterial isolates showed green metallic sheen while 4 (10%) isolates showed no growth on EMB agar. Another similar study indicates the high prevalence of E.coli in meat samples [20]. Off-white colonies from MacConkey agar were streaked on SS agar for further identification of bacterial isolates. It was observed that in chicken samples, more bacterial isolates (78%) showed black colonies on SS agar as compared to beef and mutton while in mutton samples, more bacterial isolates (22%) showed colorless colonies on SS agar.
Gram-positive cocci were streaked on mannitol salt agar to check the ability of bacteria to ferment mannitol. Among Gram-positive cocci 100% isolates from chicken, 70% from beef, and 50% from mutton showed yellow growth on MSA. These gram-positive isolates were checked for hemolytic activity. It was observed that about 72% of bacterial isolates showed beta hemolysis while 28% of bacterial isolates showed gamma hemolysis on blood agar. All the isolates from chicken showed beta hemolysis, 70%gram-positive cocci from beef showed beta hemolysis while gamma hemolytic isolates and gamma hemolytic isolates were equally present in mutton samples.
Biochemical tests were performed to confirm the identification of selected organisms. Citrate, oxidase, and IMViC tests were performed. A catalase test was performed on all isolates while an oxidase test was done on gram-negative bacterial isolates. IMViC tests were also performed to differentiate members of coliform. It was observed that E. coli was the more common (34%) pathogenic bacteria found in raw chicken followed by Salmonella (28%), Staphylococcus (25%), Shigella (8%), and Bacillus (3%). E. coli serves as a sanitary indicator of a slaughterhouse or poor handling while other bacteria listed above are the main causes of chicken-borne diseases [17]. In beef samples, E. coli (39%) was more common followed by Salmonella (30%), Staphylococcus (18%) and Enterobacter (8%), and Shigella (5%) while in mutton samples E. coli (32%), Salmonella (32%), Staphylococcus (12%), Shigella (12%), Enterobacter (9%) and Bacillus (3%). E. coli which can contaminate meat products are also classified in the group of coliforms and fecal coliforms [18]. Prevalence of E. coli in meat samples might be due to the contact of fecal material or intestinal part with the carcass during the slaughter and there is a high chance of the mixing of E. coli during tenderization of the meat mechanically than during simply cutting the meat [19]. There are different sources of these pathogenic bacteria like the gut of animals or birds, hands of workers, biofilms on surfaces, air, and water, etc. These bacteria penetrate the meat muscle after being slaughtered [21, 22].
The antimicrobial activity of ginger was assessed against different isolates at different concentrations. It was observed that ginger shows maximum antibacterial activity against E. coli, Staphylococcus, Salmonella, Shigella, Bacillus, and Enterobacter at 0.8g/ml concentration. A mini-review conducted by Wail and Emad also indicates the antibacterial activity of ginger against various Gram-positive and Gram-negative bacteria. In this study, it was also accessed that ginger from the various geographical area shows different antibacterial activities as bio-active components present in ginger are also different in different geographical areas [23]. Different bioactive components present in ginger justify the inconsistency of present research with previous research [24, 25]. It was observed that garlic shows maximum antibacterial activity against E. coli, Staphylococcus, Salmonella, Shigella, Bacillus, and Enterobacter at 0.8g/ml concentration. Another study reported the antibacterial activity of garlic against bacteria commonly found in meat samples [26]. Maximum antibacterial activity at higher concentrations of preservatives was reported in previous studies [27, 28]. The antimicrobial activity of radish was also assessed on different isolates at different concentrations. It was observed that radish shows no antibacterial activity against E. coli, Staphylococcus, Salmonella, Shigella, Enterobacter, and Bacillus at any concentration. While a previous study indicated the antibacterial activity of radish against different bacterial strains [29]. However, results may vary according to bioactive compounds present in radish.
Antibacterial activity of synthetic preservative was checked at different concentration of sodium nitrite (0.2mM, 0.4mM, 0.6mM and 0.8mM). It was observed that sodium nitrite shows maximum inhibitory effect against E. coli, Staphylococcus, Salmonella, Shigella, Bacillus, and Enterobacter at 0.8mM concentration. Research conducted to check the antibacterial activity of different synthetic preservatives indicated that sodium nitrite is not effective in the case of certain organisms like Staphylococcus [30]. Therefore, there is a need for more effective preservatives instead of synthetic preservatives.
CONCLUSION
Ginger and garlic are commonly used spices for taste-enhancement of meat and meat products. The medicinal importance of ginger, garlic, and radish is well-known. In the presented work, the antibacterial activity of natural preservatives is observed. It was observed that ginger and garlic showed significant antibacterial activity against gram-positive and gram-negative bacteria. It can be concluded that the use of ginger and garlic as a preservative will increase the shelf-life of meat and meat products with no harmful effects. However, more research is required for the implementation of natural preservatives for food storage and safety.
References
[1] Bantawa, K., et al., Food-borne bacterial pathogens in marketed raw meat of Dharan, eastern Nepal. BMC research notes, 2018. vol 11 no 1, pp. 1-5. DOI: 10.1186/s13104-018-3722-x
[2] Tessari, P., A. Lante, and G. Mosca, Essential amino acids: master regulators of nutrition and environmental footprint? Scientific reports, 2016. vol 6 no 1, pp 1-13.DOI: 10.1038/srep26074
[3] Ishaq, A., et al., Prospect of microbial food borne diseases in Pakistan: a review. Brazilian Journal of Biology, 2021. vol 81 no 4. pp. 940-953.DOI: 10.1590/1519-6984.232466
[4] Almashhadany, D.A., Meat Borne Diseases. 2021.
http://dx.doi.org/10.1039/C7RA00172J%0Ahttps://www.intechopen.com/books/advanced-biometric-technologies/liveness-detection-in-biometrics%0Ahttp://dx.doi.org/10.1016/j.colsurfa.2011.12.014
[5] Sharaf, E.M. and S.M. Sabra, Microbiological loads for some types of cooked chicken meat products at Al-Taif Governorate, KSA. World Applied Sciences Journal, 2012. vol 17 no 5, pp. 593-597.
[6] Samad, A., et al., Multiplex polymerase chain reaction detection of Shiga toxin genes and antibiotic sensitivity of Escherichia coli O157: H7 isolated from beef meat in Quetta, Pakistan. Journal of Food Safety, 2018. vol 38 no 6, pp. e12540.DOI: 10.1111/JFS.12540
[7] Jia, W., et al., Effect of nisin and potassium sorbate additions on lipids and nutritional quality of Tan sheep meat. Food Chemistry, 2021. vol 365, pp. 130-535.
DOI: 10.1016/J.FOODCHEM.2021.130535
[8] Aminzare, M., et al., Using natural antioxidants in meat and meat products as preservatives: A review. Advances in Animal and Veterinary Sciences, 2019. vol 7 no 5, pp. 417-426. DOI: 10.17582/journal.aavs/2019/7.5.417.426
[9] Yu, H.H., Y.-W. Chin, and H.-D. Paik, Application of Natural Preservatives for Meat and Meat Products against Food-Borne Pathogens and Spoilage Bacteria: A Review. Foods, 2021. vol 10 no 10, pp. 2418. DOI: 10.3390/foods10102418
[10] Sani, M.A., A. Ehsani, and M. Hashemi, Whey protein isolate/cellulose nanofibre/TiO2 nanoparticle/rosemary essential oil nanocomposite film: Its effect on microbial and sensory quality of lamb meat and growth of common foodborne pathogenic bacteria during refrigeration. International journal of food microbiology, 2017. vol 251, pp. 8-14. DOI: 10.1016/J.IJFOODMICRO.2017.03.018
[11] Stoica, M., et al., New Strategies for the Total/Partial Replacement of Conventional Sodium Nitrite in Meat Products: a Review. Food and Bioprocess Technology, 2022 vol 15 no 3, pp. 514-538. DOI: 10.1007/S11947-021-02744-6
[12] Soni, K.A., et al., Identification of ground beef–derived fatty acid inhibitors of autoinducer-2–based cell signaling. Journal of food protection, 2008. vol 71 no 1, pp. 134-138. DOI: DOI: 10.4315/0362-028x-71.1.134
[13] Garrity, G., Bergey's Manual® of Systematic Bacteriology: Volume 2: The Proteobacteria, Part B: The Gammaproteobacteria. Vol. 2. 2007: Springer Science & Business Media. https://link.springer.com/book/10.1007/0-387-28022-7
[14] Shende, S., et al., Green synthesis of copper nanoparticles by Citrus medica Linn.(Idilimbu) juice and its antimicrobial activity. World Journal of Microbiology and Biotechnology, 2015. vol 31 no 6, pp. 865-873. DOI: DOI: 10.1007/s11274-015-1840-3
[15] Fegan, N. and I. Jenson, The role of meat in foodborne disease: Is there a coming revolution in risk assessment and management? Meat science, 2018. vol 144, pp 22-29. DOI: 10.1016/j.meatsci.2018.04.018
[16] KUMARI, M., A. KUMAR, and R. GUPTA, ENUMERATION OF TOTAL VIABLE AND TOTAL COLIFORM COUNT IN CHICKEN.2019. vol 57 no 2, pp 204-206
[17] Odeyemi, O.A., et al., Understanding spoilage microbial community and spoilage mechanisms in foods of animal origin. Comprehensive reviews in food science and food safety, 2020. vol 19 no 2,pp 311-331. https://doi.org/10.1111/1541-4337.12526
[18] Zafar, A., et al., Microbiological Evaluation of Raw Meat Products Available in Local Markets of Karachi, Pakistan: Microbial Evaluation of Raw Meat Products. Proceedings of the PakistanAcademy of Sciences: B. Life and Environmental Sciences, 2016. vol 53 no 2, pp 103-106.
[19] Nagarajan, V., et al., Study of bacterial contamination of raw meat in Hyderabad. MOJ Proteomics Bioinform, 2018. vol 7 no 1, pp. 46-51. DOI: 10.15406/mojpb.2018.07.00212
[20] Tassew, H., et al., Microbial flora and food borne pathogens on minced meat and their susceptibility to antimicrobial agents. Ethiopian journal of health sciences, 2010. vol 20 no 3, DOI: 10.4314/ejhs.v20i3.69442
[21] Rouger, A., O. Tresse, and M. Zagorec, Bacterial contaminants of poultry meat: sources, species, and dynamics. Microorganisms, 2017. vol 5 no 3 pp 50. doi: 10.3390/microorganisms5030050
[22] Bakhtiary, F., et al., Evaluation of bacterial contamination sources in meat production line. Journal of food quality, 2016. vol 39 no 6, pp. 750-756. https://doi.org/10.1111/jfq.12243
[23] Abdalla, W.E. and E.M. Abdallah, Antibacterial activity of ginger (Zingiber Officinale Rosc.) Rhizome: a mini review. Int J Pharmacogn Chinese Med, 2018. vol 2, pp. 1-142.
[24] Yassen, D. and A.E. Ibrahim, Antibacterial activity of crude extracts of ginger (Zingiber officinale Roscoe) on Escherichia coli and Staphylococcus aureus: A Study in vitro. Indo American Journal of Pharmaceutical Research, 2016. vol 6 no 6, pp. 5830-5835.
[25] Kadogo, J., Evaluation of antimicrobial activity of ginger and garlic extracts against isolates of Staphylococcus aureus, Escherichia coli and Salmonella species. 2021, Makerere University. http://hdl.handle.net/20.500.12281/9447
[26] Tareq, M., S. Rahman, and M. Hashem, Effect of clove powder and garlic paste on quality and safety of raw chicken meat at refrigerated storage. World J Nutr Food Sci, 2018. vol 1 no 1, p. 1002
[27] Karuppiah, P. and S. Rajaram, Antibacterial effect of Allium sativum cloves and Zingiber officinale rhizomes against multiple-drug resistant clinical pathogens. Asian Pacific journal of tropical biomedicine, 2012. vol 2 no 8, pp. 597-601. doi: 10.1016/S2221-1691(12)60104-X
[28] Sikrodia, R., et al., Efficacy of Antimicrobial Activity of Aqueous Extract of Garlic (Allium sativum) and Ginger (Zingiber officinale) against Different Bacterial Species. Int. J. Curr. Microbiol. App. Sci, 2018. vol 7 no 9, pp. 3021-3025.
[29] Kaymak, H.C., et al., In vitro antibacterial activities of black and white radishes (Raphanus sativus L.). Comptes Rend. L Acad. Bulgare Des. Sci, 2015.vol 68: pp 201-208. https://hdl.handle.net/20.500.12619/67393
[30] Lin, L., et al., Assessment of the inhibitory effects of sodium nitrite, nisin, potassium sorbate, and sodium lactate on Staphylococcus aureus growth and staphylococcal enterotoxin A production in cooked pork sausage using a predictive growth model. Food Science and Human Wellness, 2018. vol 7 no 1, pp 83-90. DOI: 10.1016/j.fshw.2017.12.003




















