Tomato Disease Classification using Fine-Tuned Convolutional Neural Network
Haseeb Younis1, Muhammad Asad Arshed2, Fawad ul Hassan3, Maryam Khurshid 4, Hadia Ghassan5
1Department of Computer Science, University College Cork, Cork, Ireland
2Department of Software Engineering, University of Management & Technology, Lahore, Pakistan
3Superior University, Lahore Campus, Pakistan
4Comsats University Islamabad, Lahore Campus, Pakistan
5Department of Computer Science, Minhaj University, Lahore, Pakistan
* Correspondence: Muhammad Haseeb– Email ID:hahhaseeb@gmail.com
Citation |Younis. H, Arshed. M. A, Hassan. F, Khurshid. M, Ghassan. H, “Tomato Disease Classification using Fine-Tuned Convolutional Neural Network”. International Journal of Innovations in Science and Technology. Vol 4, issue 1, pp: 123-134, 2022.
Received |Jan 11, 2022; Revised | Feb 5, 2022 Accepted |Feb 6, 2022; Published | Feb 13, 2022._________________________________________________________________
Tomatoes have enhanced vitamins that are necessary for mental and physical health. We use tomatoes in our daily life. The global agricultural industry is dominated by vegetables. Farmers typically suffer a significant loss when tomato plants are affected by multiple diseases. Diagnosis of tomato diseases at an early stage can help address this deficit. It is difficult to classify the attacking disease due to its range of manifestations. We can use deep learning models to identify diseased plants at an initial stage and take appropriate measures to minimize loss through early detection. For the initial diagnosis and classification of diseased plants, an effective deep learning model has been proposed in this paper. Our deep learning-based pre-trained model has been tuned twofold using a specific dataset. The dataset includes tomato plant images that show diseased and healthy tomato plants. In our classification, we intend to label each plant with the name of the disease or healthy that is afflicting it. With 98.93% accuracy, we were able to achieve astounding results using the transfer learning method on this dataset of tomato plants. Based on our understanding, this model appears to be lighter than other advanced models with such considerable results and which employ ten classes of tomatoes. This deep learning application is usable in reality to detect plant diseases.
Keywords: Classification of Disease Tomato Plants, Mobile-Net, Plant Disease Classification, Transfer Learning
INTRODUCTION
A new era of a massive economy and a large population production of food is required to feed humans. However, diseases in plants cater to the need of severe loss for healthy farmers and everybody else. Globally, these diseases have caused the loss of 21.5 % wheat, 30% rice, 21.4% soybean, 22.5% maize, and 17.2% potato in 2019 [1]. In Punjab and Sindh, a disease of Tomato leaf curl has resulted in the crop loss of 30% - 40%. This led to severe economic losses and the need to import tomatoes at exorbitant prices [2], [3]. Using images to classify plant diseases is a challenging task with several factors to consider, including the presence of symptoms accompanied by different appearances, the possibility that different illnesses could cohabit in a significant plant, and a multitude of disorders that share similar symptoms [4]. Factors like image background inferences as well as variations in lighting encountered during data capturing are further complicated. The classifier trained on the images must be given a major representation of the features for traditional image classification. Color, shape, or texture may be traits that distinguish a plant disease but identifying them and categorizing an image based on these traits is difficult. [5].
Due to the issues of traditional machine learning, researchers are taking interest in deep learning based convolutional neural networks. The abstraction power of CNN layers allows to extricate high-level (texture and semantics) and low-level features (shapes and edges) automatically. This was shown in recent research [6].
For the identification, segmentation, classification and data generation of images from various domains, a number of classifications have been used by researchers such as machine learning (ML), computer vision (CV), image recognition, and deep learning techniques. [7] [8]. To classify herb plants and detect their early diseases, an automated hybrid smart system was employed by M.S.Mustafa et al. [9] that can classify hard plants accurately. An extensive process of identifying plants manually relied on knowledge of the plant itself such as texture, odor, and shape. The objective of the proposed study was to elucidate the identification and detection of organisms and early herb plant diseases through the significance of these functions. For the identification of early herb disease can be identified by leaves' color, smell, shape, and texture, a method. This method was based on a hybrid intelligent system that involved a probabilistic neural network, SVM classifier fuzzy, Naive Bayes and inference system. It also classified species by utilizing computer vision and electronic noses. [10].
The researcher Barbedo et al. [11] showed how the size of the dataset impacted the efficiency of deep learning and shift learning for plant disease classification. Conventional machine learning approaches such as Multilayer Perceptron Neural Networks (MPNN), Decision Trees, and SVM were employed to study automated identification of plant diseases. Despite this, there has been a shift toward the implementation of deep learning methods based on convolutional neural networks (CNN). To operate properly, these approaches used a large number of datasets that consisted of a wide range of situations. These limitations were considered significant when an adequate database of images was developed. Through deep learning approaches, explored how the size and complexity of datasets could influence the accuracy of plant pathology. The researcher used a database of images showing twelve different plant types in their study [11]. Each plant type contained unique characteristics concerning the intensity of disease, range of circumstances, and the number of samples. Results of the experimental study showed that most of the technical limitations were controlled, which were linked with automatic classification of plant disease but, the data used for training purposes had negative implications and hindered the proliferation of these technologies.
De Chant et al. [12] employed an automated detection system by implementing deep learning to detect diseased maize plants with northern leaf blight from field imaging. The proposed system recognized whether a disease existed or not in an image. In Particular, disease occurrence was approximated by this information.
The methodology of the study involved the (CNN) framework to overcome the issues associated with minimal data and numerous irregularities that emerged in images of field-grown plants. The main limitation of their detection system was to conduct manual identification of those images that were trained by using CNN. On the set of images that were not in training, the system achieved an accuracy rate of 96.7% [12].
The features that were automatically extracted by these CNN's performed well. However, a drawback of CNN was that massive training data is required as input which makes it data-hungry. To solve this problem, transfer learning (a technique in which a pre-trained model is applied) was used [13]. Pre-trained models were also used by various researchers for the plant disease classification. A deep learning-based detector was employed by Alvaro Fuentes et al. [14] to determine real-time disease in tomato plants and seeds. Single-Shot Multibox Detector (SSD), Region-Based Fully Convolutional Network (R-FCN), and Faster Region-based Convolutional Neural Network (Faster R-CNN) are three detectors that were used in their study. and it was presented as "Deep learning meta-architectures". The 'deep features extractors' like ResNet (Residual Network) and VGG net were combined with these meta deep learning architectures. For the classification of plant disease, the convolutional architectures were presented by Kamal KC et al. [15] [3]. These convolutional architectures were depth-wise separable and their two kinds of building blocks were used by researchers to present two versions. They also executed training and testing on a PlantVillage dataset that is publicly accessible. This dataset contained 55 various classes and 82,161 images to discriminate between healthy and diseased plants. A high rise in convergence speed and a low precision were attained by these depth-wise separable convolutions. Reduced MobileNet achieved a classification accuracy of 98.34% compared to other training and testing models.
VGG16 [17] (a CNN model that is proposed by K. Simonyan and A. Zisserman in paper “Very Deep Convolutional Networks for Large-Scale Image Recognition) was used by D.oppenheim et al. [16] with deep learning, which greatly enhanced the capabilities of computer vision (CV), such as appearances and current developments. It was possible to identify and classify particular plant diseases accurately based on certain visually discernible signs. To classify tubers (specialized storage stems of certain seed plants)) into four groups based on their diseases, a balanced potato class, and five categories, a deep CNN was used by the algorithm [16]. Correspondingly, Alexnet [19] has been used for the identification of rice disease by Yang LU et al. [18]. Ten distinct diseases can be classified by this method. The dataset that was obtained from experimental rice fields was used by researchers and it contained 500 natural images of healthy and diseased stems and leaves of rice. By the implementation of the validation approach of the ten cross-fold, their proposed experimental model of CNN attained an accuracy of 95.48%. Simulated results were used to establish the efficacy and feasibility of the proposed approach for identifying rice diseases. Table 1 shows the comparison between these studies.
A multiclass classification model of tomato plant disease was employed in this paper. Our primary objective was to classify various plant diseases. In this study, MobileNet CNN [22] was fine-tuned on 1.3 million pre-trained images. This network was applied to the dataset of tomato plant disease classification. We collected this dataset and it consisted of 10 distinct classes, which contained 18,160 images. Healthy Early blight, Late blight, Bacterial spot, Septoria leaf spot, Spider_mites Two-spotted, Target_Spot, Leaf Mold, Yellow Leaf Curl Virus, and mosaic virus are the classes involved in this dataset. Our model attained an accuracy of 98.93%. In section 3, our proposed work is described in detail containing thorough explanations of the dataset and results.
Table 1. Comparison of various state-of-the-arts studies
|
Studies |
Dataset |
Methods |
Classes |
Accuracy % |
|
Barbedo et al [11] (2018) |
12 plants |
GoogleNet |
56 |
84% |
|
De Chant et al [12] (2017) |
Corn Images |
Pipeline |
2 |
97 % |
|
Fuentes et al [14] (2017) |
Tomato images |
Multiple networks |
10 |
83% |
|
KC et al [15] (2019) |
Plant Village |
MobileNet |
55 |
98% |
|
Oppenheim et al [16] (2017) |
Potato Images |
VGG |
5 |
96% |
|
LU et al [18] (2018) |
Rice Images |
AlexNet |
10 |
95% |
|
Sinha et al [20] (2020) |
Plant Village |
Pipeline |
55 |
74% |
|
Chen et al [21] (2021) |
Crop Disease (own) |
CNN |
4 |
93.75% |
MATERIALS & METHODS
We applied the following methodology for creating the multiclass disease of tomato plants, classification model. During the experimental phase, RAM of 2 GB, Core i7 System 7800 Model as well as GTX 1080-Ti were used. For the evaluation of the proposed model, we considered accuracy as well as loss curves for evaluation and compared them to literature studies. The flow of study is shown in Figure 1.
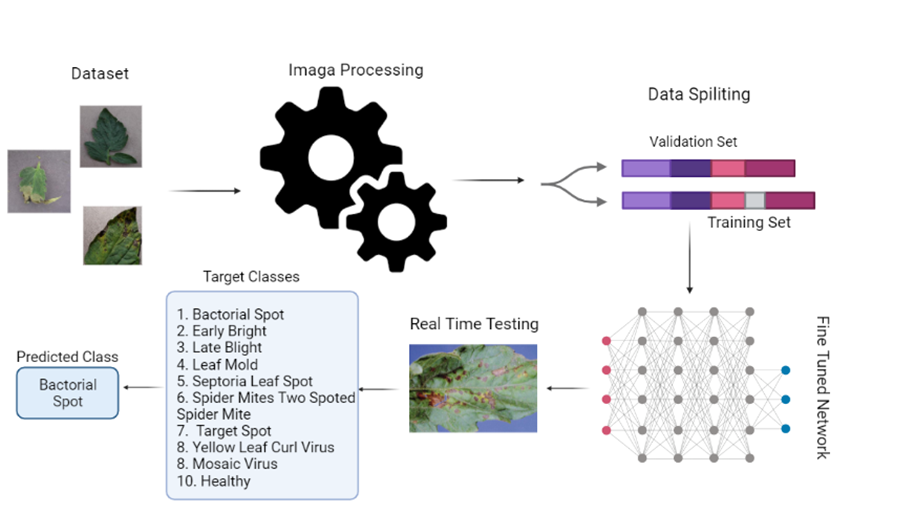
Figure 1. Abstract Diagram
Dataset Overview: This study used the dataset of tomato disease classification that showed the ten most common diseases of tomato. In table 2, a comprehensive portrayal of several images and names of their classes has been shown. These images are present in the dataset to train and validate the data[1].
Table 2. Dataset Classes and no. of images
|
Classes |
Train Images |
Validation Images |
Total Images |
|
Tomato Bacterial spot |
1000 |
100 |
1100 |
|
Tomato Early blight |
1000 |
100 |
1100 |
|
Tomato Late blight |
1000 |
100 |
1100 |
|
Tomato Leaf Mold |
1000 |
100 |
1100 |
|
Tomato Septoria leaf spot |
1000 |
100 |
1100 |
|
Tomato Spider Mites Two spotted spider mite |
1000 |
100 |
1100 |
|
Tomato Target Spot |
1000 |
100 |
1100 |
|
Tomato Yellow Leaf Curl Virus |
1000 |
100 |
1100 |
|
Tomato mosaic virus |
1000 |
100 |
1100 |
|
Tomato healthy |
1000 |
100 |
1100 |
|
Total |
10000 |
1000 |
11000 |
Data Preprocessing: Ten distinct classes containing 11,000 images have been included in this dataset, which contains images of affected plants. These classes are as follows: Healthy Early blight, Late blight, Bacterial spot, Septoria leaf spot, Spider_mites Two-spotted, Target_Spot, Leaf Mold, Yellow Leaf Curl Virus, and mosaic virus affected plants images. Data was divided into three groups for experimental purposes: training, validation, and testing. Initially training images contained 10,000 images, 1000 images for validation, and the remaining images were obtained from the internet for real-time testing. During training, we used the original number of images, however, the batches are made up of size 32. At the end of the training, we took 92 batches for validation purposes and 22 to test the model. Moreover, for our model's better efficiency, we implemented 0.2 random rotation and random flip which is horizontal to perform augmentation. Although images were altered to various angles and positions, a better classification of data was attained by this data augmentation. Figure 2 shows the sample images of considered dataset.
 Figure 2. Sample Images [2] for Augmentation of Data using 0.2 random rotation and horizontal Random Flip
Figure 2. Sample Images [2] for Augmentation of Data using 0.2 random rotation and horizontal Random Flip
Fine-Tuning and Training of Models: In this dataset, we utilized two-fold of the model which is pre-trained MobileNet for training the disease classifier. For the initial fold, the weights of pre-trained ImageNet of the MobileNet were imported and then the layers were frozen that were trainable. The feature batch of the dataset in the training section was passed to the 2D global average pooling layer after we added the revert input layers of MobileNet V2. Our final step was to feed the inputs to the augmentation layers, which were built from the base model and the input layer. After dropping out of 0.2, here was also a global average layer adjacent to it. The 2nd fold consisted of model fine-tuning as well as enabling a trainable section from the model that was imported. Originally, the base model contained 155 sections of layers, however, starting with the layers, they were fine-tuned in the range of 100-155 and 100 layers before that were removed. In figure 3, two model’s architectures have been shown and table 3 displays hyper parameters of these two folds.
Table 3. Tuned Parameters of considered methodology
|
Model Name |
Optimizer (Algorithm) |
LR-Learning Rate |
Metric |
Loss-Function |
Epochs |
|
Mobile Net |
Adam |
0.0001 |
Accuracy |
Sparse Categorical Cross-entropy |
50 |
|
Tuned Mobile Net |
RMSprop |
0.0001/10 |
Accuracy |
Sparse Categorical Cross-entropy |
50 |
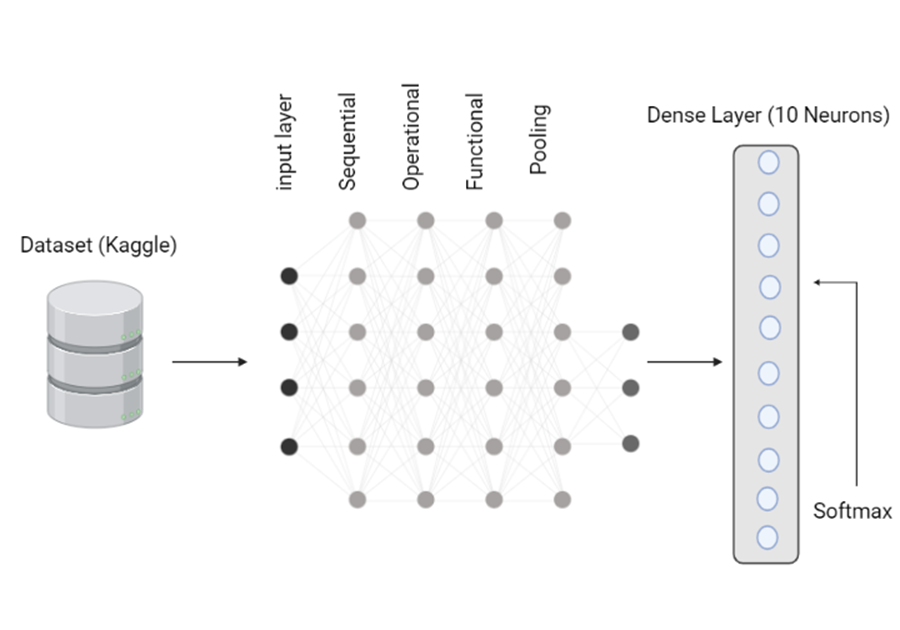
Figure 3. Proposed Model Architecture
For the evaluation measure, the overall accuracy was examined using equation 1.
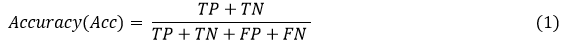
Where TP represents the recall or truly predicted image, True Negative is directed by TN, FP demonstrates the false positive, and FN displays the false negative.
RESULTS
In this study, transfer learning technique that is application of deep learning is considered for classification of objects. The augmented images are passed to the model. The last layer of the network in deep learning is equal to the number of classes. In our study, we have a total of 10 classes, from which 9 are of tomato disease and 1 represents health plants. Further, we have considered dropout techniques to avoid overfitting of the network. For getting 1 label from 10, we have applied softmax in the last layer of the network. For the performance calculation of the suggested model, loss and accuracy was used, which resulted in achieving a loss rate of 0.3788 and validation accuracy of 86.98%, in the first fold.
Dropout: To avoid the overfitting, dropout rate plays an important role in deep learning models as well as batch size and epoch. We have set the dropout layers between dense layers to overcome the overfitting problem. The training and validation accuracy graph lines should be close to each other to demonstrate that the model is effective, it has no overfitting. We have 50 epochs, which is also not too much. Due to all these parameters, we are able to achieve noticeable results. All the parameters are figured out from a huge list and we have set the best parameters in our study for effective results e.g., epochs 50, batch size 32 and optimizer RMSprop.
Training & Validation Accuracy without Fine Tuning Approach: Accuracy is a metric that is mainly used for classification problems. It is a percentage that determines how much your test data is correctly classified. For example, you have 100 samples as validation, and from these samples 90 are correctly classified that means the proposed model is 90% effective. From training and validation accuracy, the most important metrics is validation accuracy, because our purpose of model is to design a model that performs effectively for unseen data. Without applying a fine-tuning approach, the results of our proposed model are sown in Figure 4.
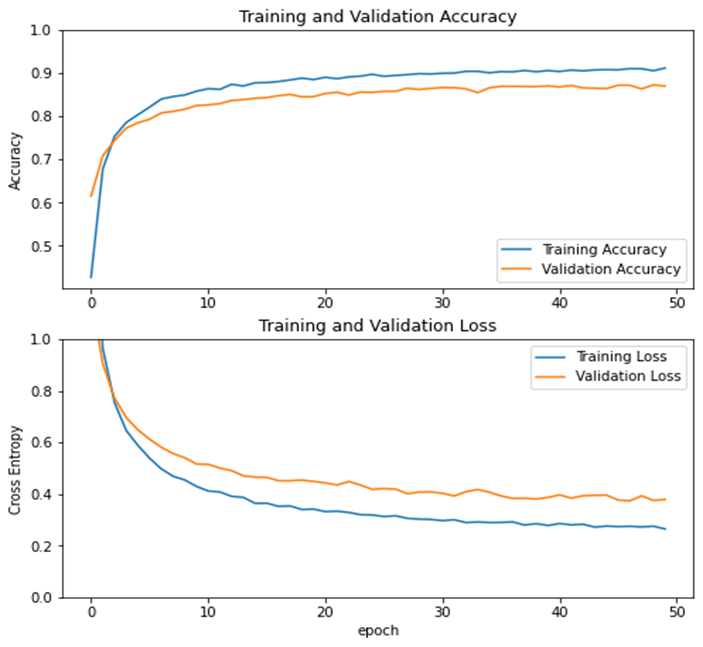
Figure 4. Learning graph accuracy as well as loss without fine tuning
Training & Validation Accuracy with Fine Tuning Approach: Fine tuning is an effective approach that means a small adjustment in the process to achieve the desired output. In the fine-tuning approach, we used weights of the previous model to design a similar network. Weights are connections between neurons of a deep learning model. We have used this approach in this study to save our time to design a model from scratch as this method already has some information. A well-known fine-tuning framework are Keras, Torch and Tensor Flow. The base model was Mobile Net and we have tuned it with a fine-tuning approach and Figure 5 illustrates that fine tune approach is effective and we are able to achieve the validation accuracy of 98.93% that is roughly 11.95 % greater than normal mobile net method validation accuracy 86.98%.

Figure 5. Learning graph accuracy as well as loss with fine tuning
Discussion
We have proposed an effective model for tomato disease. For the robustness of the proposed model, initially we have compared our results to existing studies in terms of the number of parameters (that is an important part to design a light weight model).
- The total number of parameters in our model is 2.3 million, which is very low. Among 2.3 million parameters, only 48,578 were trainable for the first model.
- Oppenheim et al. [16] proposed a model containing parameters of 14.7 million.
- LU et al. [18] (2018) proposed model contained parameters of 3.3 million.
- KC et al. [15] proposed model has 3.2 million parameters.
Further we have enhanced our model with a fine tune approach due to which the proposed model has now 1.9 million trainable parameters, which is less than all comparable studies and illustrates our model is lightweight. Figure 6 illustrates the effectiveness of fine tune based proposed model, that illustrate that our model is effective than other 8 well known models.
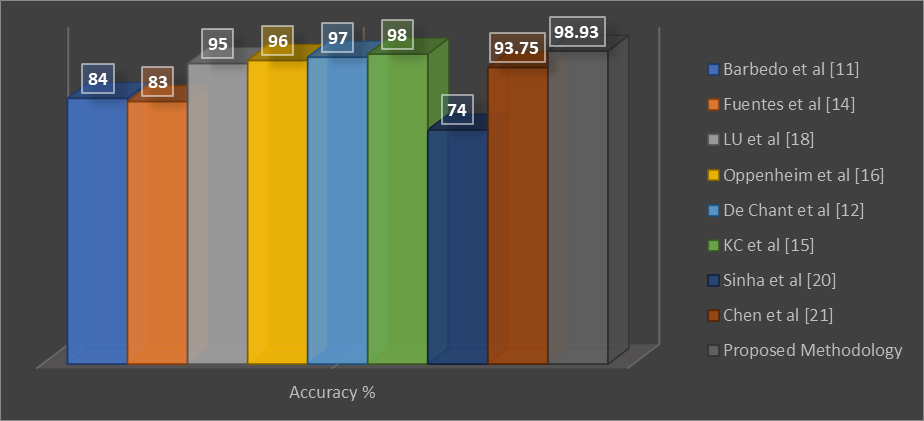
Figure 6. Mean Evaluation Scores of Different Models
CONCLUSION
To maintain the economic and ecological well-being of plants, early detection of plant disease needs time. With only the naked eye, it is difficult for the manufacturers to detect the disease which is causing the plant to be destroyed. A system that can help farmers is needed. Therefore, in our study, a lightweight multiclass plant disease classification model has been proposed with significant accuracy based on a pre-trained Mobile Net CNN. A dataset we collected containing ten classes of tomato plants was used to train the model. Both healthy and diseased plants’ images are included in our dataset. By using our methodology, we achieved 98.93% accuracy, which is better than other comparative studies. We have developed a lightweight model that can be conveniently deployed through smart mobile applications. It can instantly classify the plants.
Author’s Contribution. All the authors contributed equally.
Conflict of interest. We declare no conflict of interest for publishing this manuscript in IJIST.
REFERENCES
[1] S. Savary, L. Willocquet, S. J. Pethybridge, P. Esker, N. McRoberts, and A. Nelson, "The global burden of pathogens and pests on major food crops," Nat. Ecol. Evol., vol. 3, no. 3, pp. 430–439, Mar. 2019.
[2] S. Mansoor et al., "Evidence for the association of a bipartite geminivirus with tomato leaf curl disease in Pakistan," Plant Disease, vol. 81, no. 8. Plant Dis, p. 958, 1997.
[3] A. Raza et al., "First Report of Tomato Chlorosis Virus Infecting Tomato in Pakistan," Plant Dis., vol. 104, no. 7, pp. 2036–2036, Jul. 2020.
[4] J. G. A. Barbedo, "A review on the main challenges in automatic plant disease identification based on visible range images," Biosystems Engineering, vol. 144. Academic Press, pp. 52–60, 01-Apr-2016.
[5] A. García-Floriano, Á. Ferreira-Santiago, O. Camacho-Nieto, and C. Yáñez-Márquez, “A machine learning approach to medical image classification: Detecting age-related macular degeneration in fundus images,” Comput. Electr. Eng., vol. 75, pp. 218–229, May 2019.
[6] W. Zhao and S. Du, "Spectral-Spatial Feature Extraction for Hyperspectral Image Classification: A Dimension Reduction and Deep Learning Approach," IEEE Trans. Geosci. Remote Sens., vol. 54, no. 8, pp. 4544–4554, Aug. 2016.
[7] M. Z. Khan et al., "A Realistic Image Generation of Face from Text Description Using the Fully Trained Generative Adversarial Networks," IEEE Access, vol. 9, pp. 1250–1260, 2021.
[8] G. Khan, S. Jabeen, M. Z. Khan, M. U. G. Khan, and R. Iqbal, "Blockchain-enabled deep semantic video-to-video summarization for IoT devices," Comput. Electr. Eng., vol. 81, p. 106524, Jan. 2020.
[9] M. S. Mustafa, Z. Husin, W. K. Tan, M. F. Mavi, and R. S. M. Farook, "Development of an automated hybrid intelligent system for herbs plant classification and early herbs plant disease detection," Neural Comput. Appl., vol. 32, no. 15, pp. 11419–11441, Aug. 2020.
[10] M. S. Mustafa, Z. Husin, W. K. Tan, M. F. Mavi, and R. S. M. Farook, "Development of an automated hybrid intelligent system for herbs plant classification and early herbs plant disease detection," Neural Comput. Appl., vol. 32, no. 15, pp. 11419–11441, Aug. 2020.
[11] J. G. A. Barbedo, "Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification," Comput. Electron. Agric., vol. 153, pp. 46–53, Oct. 2018.
[12] C. DeChant et al., "Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning," Phytopathology, vol. 107, no. 11, pp. 1426–1432, Nov. 2017.
[13] H. Younis, M. H. Bhatti, and M. Azeem, "Classification of skin cancer dermoscopy images using transfer learning," in 15th International Conference on Emerging Technologies, ICET 2019, 2019.
[14] A. Fuentes, S. Yoon, S. Kim, and D. Park, "A Robust Deep-Learning-Based Detector for Real-Time Tomato Plant Diseases and Pests Recognition," Sensors, vol. 17, no. 9, p. 2022, Sep. 2017.
[15] K. KC, Z. Yin, M. Wu, and Z. Wu, "Depthwise separable convolution architectures for plant disease classification," Comput. Electron. Agric., vol. 165, p. 104948, Oct. 2019.
[16] D. Oppenheim and G. Shani, "Potato Disease Classification Using Convolution Neural Networks," 2017.
[17] H. Qassim, A. Verma, and D. Feinzimer, "Compressed residual-VGG16 CNN model for big data places image recognition," in 2018 IEEE 8th Annual Computing and Communication Workshop and Conference, CCWC 2018, 2018, vol. 2018-January, pp. 169–175.
[18] Y. Lu, S. Yi, N. Zeng, Y. Liu, and Y. Zhang, "Identification of rice diseases using deep convolutional neural networks," Neurocomputing, vol. 267, pp. 378–384, Dec. 2017.
[19] M. Z. Alom et al., "The History Began from AlexNet: A Comprehensive Survey on Deep Learning Approaches," arXiv, Mar. 2018.
[20] A. Sinha and R. Singh Shekhawat, "A novel image classification technique for spot and blight diseases in plant leaves," Imaging Sci. J., vol. 68, no. 4, pp. 225–239, May 2020.
[21] J. Chen, J. Chen, D. Zhang, Y. A. Nanehkaran, and Y. Sun, "A cognitive vision method for the detection of plant disease images," Mach. Vis. Appl., vol. 32, no. 1, p. 3, Feb. 2021.
[22] A. G. Howard et al., “MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications,” arXiv, Apr. 2017.
[1]https://www.kaggle.com/kaustubhb999/tomatoleaf
[2]https://www.kaggle.com/kaustubhb999/tomatoleaf
 |
Copyright © by authors and 50Sea. This work is licensed under Creative Commons Attribution 4.0 International License. |




















