Towards Skin Cancer Classification Using Machine Learning and Deep Learning Algorithms: A Comparison
DOI:
https://doi.org/10.33411/ijist/2021030508Keywords:
skin cancer, classification, ISIC, International Skin Imaging Collaboration, machine learning, CNN, LR, NB, DT, KNNAbstract
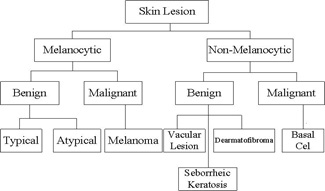
Skin cancer is an uncontrolled development of abnormal skin cells potentially due to excessive exposure to sun, history of sunburns, less melanin, Precancerous skin lesions, moles, etc. This occur when unrepaired DNA damages the cells of the skin. It is one of the diseases that are viewed on its quick evolution and the most common type of cancer that endangers life. Researchers have implemented several machine learning and deep learning techniques for classification of skin cancer. In this research paper, different cancer categories are classified using significant attributes. We have used International Skin Imaging Collaboration (ISIC) dataset for classification purposes. This dermoscopic attributes dataset includes 1000 images and 10016 instances, seven categories, 5 features and 2 Meta attributes. We implemented K-Nearest Neighbor, Logistic Regression, Convolutional Neural Network, Naïve Bayes, and Decision Tree for classification and compared their performance. In order to implement classification algorithm, we used Orange which is an open-source machine learning, data mining, and data visualization toolkit. The models are evaluated based on matrices that include Accuracy, C. Automation, F1 score, Precision, Recall, and AUC. Furthermore, frequency of features is visualized using graphical method and the ROC analysis is also performed for the classifiers. It is observed that CNN technique provided the highest accuracy of 89% and the mentioned results are the highest results of classification with the state of the art techniques. For future, the improved and recent dataset and ensemble modelling techniques based on deep learning can used to enhance classification results. The research can also be extended for other cancer types using CNN.
References
U. Jamil, A. Sajid, M. Hussain, O. Aldabbas, A. Alam, and M. U. Shafiq, “Melanoma segmentation using bio-medical image analysis for smarter mobile healthcare,” J. Ambient Intell. Humaniz. Comput., 2019, vol. 10, no. 10, pp. 4099–4120.
J. Burdick, O. Marques, J. Weinthal, and B. Furht, “Rethinking Skin Lesion Segmentation in a Convolutional Classifier,” J. Digit. Imaging, vol. 31, 2018, no. 4, pp. 435–440.
K. D. Miller et al., “Cancer treatment and survivorship statistics, 2016,” CA. Cancer J. Clin.,2016, vol. 66, no. 4, pp. 271–289.
T. Akram, M. A. Khan, M. Sharif, and M. Yasmin, “Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features,” J. Ambient Intell. Humaniz. Comput., 2018.
E. Nasr-Esfahani et al., “Melanoma detection by analysis of clinical images using convolutional neural network,” Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS, 2016, vol. 2016-Octob, pp. 1373–1376.
M. A. Khan, M. Y. Javed, M. Sharif, T. Saba, and A. Rehman, “Multi-model deep neural network-based features extraction and optimal selection approach for skin lesion classification,” 2019 Int. Conf. Comput. Inf. Sci. ICCIS 2019, 2019, pp. 1–7.
M. Properties and H. O. Mucosa, “Technical Report”, 2015, pp. 2–3.
P. Tschandl, C. Rosendahl, and H. Kittler, “Data Descriptor: The HAM 10000 dataset, a large collection of multi-sources dermatoscopic images of common pigmented skin lesions,” Nat. Publ. Gr., 2018, vol. 5, pp. 1–9.
P. Dubai, S. Bhatt, C. Joglekar, and S. Patii, “Skin cancer detection and classification,” Proc. 2017 6th Int. Conf. Electr. Eng. Informatics Sustain. Soc. Through Digit. Innov. ICEEI 2017, 2018, vol. 2017-Novem, pp. 1–6.
M. A. Farooq, M. A. M. Azhar, and R. H. Raza, “Automatic Lesion Detection System (ALDS) for Skin Cancer Classification Using SVM and Neural Classifiers,” Proc. - 2016 IEEE 16th Int. Conf. Bioinforma. Bioeng. BIBE 2016, 2016, pp. 301–308.
R. Ashraf, I. Kiran, T. Mahmood, A. Ur Rehman Butt, N. Razzaq, and Z. Farooq, “An efficient technique for skin cancer classification using deep learning,” Proc. - 2020 23rd IEEE Int. Multi-Topic Conf. INMIC 2020, 2020.
H. R. Mhaske and D. A. Phalke, “Melanoma skin cancer detection and classification based on supervised and unsupervised learning,” 2013 Int. Conf. Circuits, Control. Commun. CCUBE 2013, pp. 1–5, 2013.
K. Ramlakhan and Y. Shang, “A mobile automated skin lesion classification system,” Proc. - Int. Conf. Tools with Artif. Intell. ICTAI, 2011, pp. 138–141.
A. U. Rehman Butt, W. Ahmad, R. Ashraf, M. Asif, and S. A. Cheema, “Computer Aided Diagnosis (CAD) for Segmentation and Classification of Burnt Human skin,” 1st Int. Conf. Electr. Commun. Comput. Eng. ICECCE 2019, 2019, no. July, pp. 24–25.
C. L. Chang and C. H. Chen, “Applying decision tree and neural network to increase quality of dermatologic diagnosis,” Expert Syst. Appl., 2009, vol. 36, no. 2 PART 2, pp. 4035–4041.
R. Z. B and E. Conchon, “eHealth 360°,” , 2017, vol. 181, pp. 407–418.
A. Hekler et al., “Superior skin cancer classification by the combination of human and artificial intelligence,” Eur. J. Cancer, 2019, vol. 120, pp. 114–121.
F. A. Khan et al., “Computer-aided diagnosis for burnt skin images using deep convolutional neural network,” Multimed. Tools Appl., 2020, vol. 79, no. 45–46, pp. 34545–34568.
A. Ech-Cherif, M. Misbhauddin, and M. Ech-Cherif, “Deep Neural Network Based Mobile Dermoscopy Application for Triaging Skin Cancer Detection,” 2nd Int. Conf. Comput. Appl. Inf. Secur. ICCAIS 2019, 2019, pp. 1–6.
Z. Abbas, M. U. Rehman, S. Najam, and S. M. Danish Rizvi, “An Efficient Gray-Level Co-Occurrence Matrix (GLCM) based Approach Towards Classification of Skin Lesion,” Proc. - 2019 Amity Int. Conf. Artif. Intell. AICAI 2019, 2019, pp. 317–320.
F. A. Khan, A. U. Rehman Butt, M. Asif, H. Aljuaid, A. Adnan, S. Shaheen, “Burnt Human Skin Segmentation and Depth Classification Using Deep Convolutional Neural Network (DCNN),” J. of Medical Imaging and Health Informatics, 2020, vol. 10, no. 10, pp. 2421-2429.
A. Mahbod, G. Schaefer, C. Wang, R. Ecker, and I. Ellinger, “Skin Lesion Classification Using Hybrid Deep Neural Networks”, Institute for Pathophysiology and Allergy Research, Medical University of Vienna, Austria Department of Research and Development, TissueGnostics GmbH , Austria Department of Computer Science , Loughborough University , U . K . Department of Biomedical, 2019, pp. 1229–1233.
A. Krizhevsky, I. Sutskever, and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Commun. ACM, 2017, vol. 60, no. 6, pp. 84–90.
F. O. R. L. Arge and C. I. Mage, “V d c n l -s i r,” , 2015, pp. 1–14.
V. Sangeetha and K. J. R. Prasad, “Syntheses of novel derivatives of 2-acetylfuro[2,3-a]carbazoles, benzo[1,2-b]-1,4-thiazepino[2,3-a]carbazoles and 1-acetyloxycarbazole-2- carbaldehydes,” Indian J. Chem. - Sect. B Org. Med. Chem., 2006, 1954, vol. 45, no. 8, pp. 1951.
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 50Sea

This work is licensed under a Creative Commons Attribution 4.0 International License.




















