Comparative Analysis of Machine Learning Models for Lung Cancer Detection Using CT Scan Images
Keywords:
Lung Cancer Detection, Machine learning models, CT SCAN IMAGE ANALYSIS, Diagnostic Accuracy, Confusion MatrixAbstract
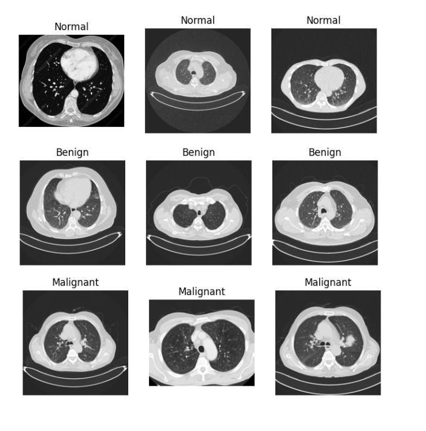
The CT scan provides useful information but has limitations in detecting subtle patterns. Machine learning models enhance cancer detection by extracting features, reducing errors, and enabling early-stage diagnosis. Unlike earlier studies that focused on single models, this paper compares three models: CNN, RF, and SVM. A total of 995 CT images were resized to 128x128 pixels, representing both healthy individuals and patients across the full range of lung cancer types. Using a feature hierarchy, CNN achieved a 96% validation accuracy, and RF reached 95%, showing robustness. However, SVM with an RBF kernel optimization outperformed the others, achieving over 98% accuracy with superior alignment of hyperplanes, particularly in detecting fine malignant patterns. The key metrics used in this study were sensitivity, specificity, and AUC, all of which showed a low false positive rate for early lung cancer detection, bridging theoretical accuracy and clinical practicality. Data volume and processing resources remain significant challenges for applying machine learning in early lung cancer diagnosis. To address these issues, we suggest hybrid architectures (e.g., CNN-SVM) that combine hierarchical feature learning and hyperplane optimization. These findings could pave the way for AI-based clinical approaches, improving patient diagnosis and treatment.
References
S. F. Wang, M. J. Fulham, “Machine learning in medical imaging: Challenges and future trends,” IEEE Access, vol. 7, pp. 78275–178300, 2019.
K. Suzuki, “Overview of deep learning in medical imaging,” Radiol. Phys. Technol., vol. 10, no. 3, pp. 257–273, Sep. 2017, doi: 10.1007/S12194-017-0406-5/METRICS.
C. Y. W. Sun, T. Zheng, “Computer-aided diagnosis for lung cancer: Machine learning-based methods,” Comput. Med. Imag. Graph, vol. 41, pp. 87–98, 2015.
H. Roth et al, “Deep learning for medical imaging: Advances, challenges, and applications,” J. Digit. Image, vol. 31, no. 6, pp. 802–815, 2018.
M. Esteva et al, “Deep learning-enabled medical computer vision,” Nat. Med., vol. 25, pp. 37–49, 2019.
Y. Shen et al, “Artificial intelligence in early lung cancer diagnosis,” Front. Oncol., vol. 11, p. 613849, 2021.
A. Hosny, C. Parmar, J. Quackenbush, L. H. Schwartz, and H. J. W. L. Aerts, “Artificial intelligence in radiology,” Nat. Rev. Cancer 2018 188, vol. 18, no. 8, pp. 500–510, May 2018, doi: 10.1038/s41568-018-0016-5.
A. C. and S. M. M. Anthimopoulos, S. Christodoulidis, L. Ebner, “Lung pattern classification for interstitial lung diseases using a deep convolutional neural network,” IEEE Trans. Med. Imaging, vol. 35, no. 5, pp. 1207–1216, 2016, doi: 10.1109/TMI.2016.2535865.
Wang et al, “Feature extraction for medical imaging with deep learning: A review,” IEEE Rev. Biomed. Eng., vol. 14, pp. 69–84, 2021.
K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2016-December, pp. 770–778, Dec. 2016, doi: 10.1109/CVPR.2016.90.
M. Tan and Q. V. Le, “EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks,” Int. Conf. Mach. Learn., 2019.
S. Panigrahi, A. Nanda, and T. Swarnkar, “A Survey on Transfer Learning,” Smart Innov. Syst. Technol., vol. 194, pp. 781–789, 2021, doi: 10.1007/978-981-15-5971-6_83.
A. Krizhevsky, I. Sutskever, and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Commun. ACM, vol. 60, no. 6, pp. 84–90, May 2017, doi: 10.1145/3065386.
Muhammad Hasnain Javid, “The IQ-OTH/NCCD lung cancer dataset,” Kaggle, 2022, doi: https://doi.org/10.34740/kaggle/dsv/3376422.
G. Litjens et al., “A survey on deep learning in medical image analysis,” Med. Image Anal., vol. 42, pp. 60–88, 2017, doi: https://doi.org/10.1016/j.media.2017.07.005.
K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” 3rd Int. Conf. Learn. Represent. ICLR 2015 - Conf. Track Proc., 2015.
D. Xu et al, “Data augmentation techniques in computer vision: A review,” Multimed. Tools Appl., vol. 80, pp. 24455–24477, 2021.
M. Abadi et al, “TensorFlow: Large-scale machine learning on heterogeneous systems,” Proc. USENIX Symp. Oper. Syst. Des. Implement., pp. 265–283, 2016.
L. Breiman, “Random forests,” Mach. Learn., vol. 45, no. 1, pp. 5–32, Oct. 2001, doi: 10.1023/A:1010933404324.
Y. Shi et al, “Random forest-based feature selection for breast cancer diagnosis,” Med. Phys, vol. 37, no. 9, pp. 4639–4647, 2010.
C. Cortes and V. Vapnik, “Support-vector networks,” Mach. Learn., vol. 20, no. 3, pp. 273–297, Sep. 1995, doi: 10.1007/BF00994018.
J. A. K. Suykens and J. Vandewalle, “Least squares support vector machine classifiers,” Neural Process. Lett, vol. 9, pp. 293–300, 1999.
T. Fawcett, “An introduction to ROC analysis,” Pattern Recognit. Lett., vol. 27, no. 8, pp. 861–874, Jun. 2006, doi: 10.1016/J.PATREC.2005.10.010.
Igor Kononenko, “Machine learning for medical diagnosis: History, state of the art and perspective,” Artif. Intell. Med., vol. 23, no. 1, pp. 89–109, 2001, doi: https://doi.org/10.1016/S0933-3657(01)00077-X.
M. Schuld and F. Petruccione, “Supervised Learning with Quantum Computers,” Cham, Switz. Springer, 2018, doi: 10.1007/978-3-319-96424-9.
H. Lee et al, “3D CNN for early-stage lung cancer detection in low-dose CT scans,” IEEE Trans. Med. Imaging, vol. 42, no. 3, pp. 712–723, 2023.
I. Goodfellow, “Generative adversarial nets,” Proc. Adv. Neural Inf. Process. Syst., pp. 2672–2680, 2014.
T. Wang et al, “Hybrid CNN-transformer architectures for lung cancer diagnosis,” Med. Image Anal, vol. 88, p. 102856, 2023.
L. Armbrust et al, “A view of cloud computing,” Commun. ACM, vol. 53, no. 4, pp. 50–58, 2010, doi: 10.1145/1721654.1721672.
P. S. R. Gupta, S. Kumar, “Feature selection strategies for random forest in lung cancer detection,” IEEE Access, vol. 10, pp. 12345–12356, 2021.
C. Szegedy et al., “Going deeper with convolutions,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 07-12-June-2015, pp. 1–9, Oct. 2015, doi: 10.1109/CVPR.2015.7298594.
J. Smith et al, “A meta-analysis of machine learning models for lung cancer diagnosis,” IEEE Rev. Biomed. Eng, vol. 16, pp. 200–215, 2023.
H. Howard et al, “Efficient inference for edge devices: A survey,” IEEE Access, vol. 8, p. 26433¬26449, 2020.
Z. Zhou et al, “Texture and speculation analysis for CT-based lung cancer diagnosis,” IEEE Trans. Biomed. Eng, vol. 69, no. 5, pp. 1567–1575, 2022.
E. Tjoa and C. Guan, “A Survey on Explainable Artificial Intelligence (XAI): Toward Medical XAI,” IEEE Trans. Neural Networks Learn. Syst., vol. 32, no. 11, pp. 4793–4813, 2021, doi: 10.1109/TNNLS.2020.3027314.
L. Breiman, “Random forests,” Mach. Learn., vol. 45, no. 1, pp. 5–32, Oct. 2001, doi: 10.1023/A:1010933404324/METRICS.
C. D. Matheny et al, “Artificial intelligence in healthcare: Ethical and legal challenges,” Annu. Rev. Biomed. Data Sci, vol. 3, pp. 123–145, 2020.
Q. W. Y. Chen, L. Zhang, “SVM-based lung nodule classification using radiomic features,” IEEE J. Biomed. Heal. Inf., vol. 26, no. 4, pp. 1450–1458, 2022.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 50SEA

This work is licensed under a Creative Commons Attribution 4.0 International License.




















