On The Crosstalk of Circadian Rhythm and Th17 cells: An Integrated Biological Regulatory Pathway
Keywords:
CD4+ Th17 cell, Cell-mediated immunity, Circadian rhythm, ROR, NFIL3Abstract
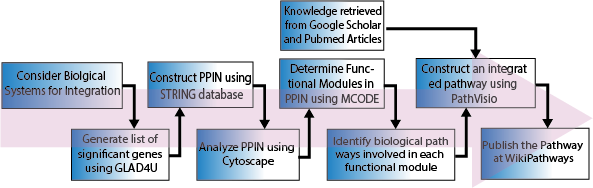
Th17 cells play a pivotal role in cell-mediated immunity and also have implications for autoimmune disorders. The interplay between the circadian rhythm and the immune system has driven interest in developing novel therapies. Th17 cells have a robust relationship with the circadian rhythm through clock-controlled genes such as NFIL3 (E4BP4), RORA, RORB, NR3C1, and RORC. The purpose of this study is to construct a literature-curated updated biological regulatory network (BRN) of the molecular regulators of circadian rhythm and CD4+ Th17 cells. The integrated BRN will provide a holistic view of the differentiation process of Th17 cells from a circadian rhythm perspective, which will enhance our understanding of the interplay between the two systems. We aim to perform formal modelling and analysis of this BRN using our previously developed approaches to gain system-wide insights into various molecular expression dynamics and identify the significance of biological clocks in immunity in the future. In addition, biological pathway databases are an integral part of omics analytical workflows, and their continuous updates with the latest knowledge are crucial for gaining biological insights from such studies. Therefore, with this additional objective, we have also uploaded this pathway to WikiPathways (Database), to facilitate its use in future studies, which can be accessed via the following URL: https://classic.wikipathways.org/index.php/Pathway:WP5130. To our knowledge, this is the first study to report a literature-curated pathway of comprehensive regulatory interactions and crosstalk between Th17 cell differentiation and circadian genes.
References
S. Zhu and Y. Qian, “IL-17/IL-17 receptor system in autoimmune disease: Mechanisms and therapeutic potential,” Clin. Sci., vol. 122, no. 11, pp. 487–511, Jun. 2012, doi: 10.1042/CS20110496,.
S. M. S.-T. John J. O’Shea, “Signal transduction and Th17 cell differentiation,” Microbes Infect. / Inst. Pasteu, vol. 11, no. 5, pp. 599–611, 2009, doi: 10.1016/j.micinf.2009.04.007.
F. S. Heather R. Conti, “Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis,” J. Exp. Med., vol. 206, no. 2, 2009, doi: 10.1084/jem.20081463.
S. L. G. Reiko M. Onishi, “Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease,” Immunology, vol. 129, no. 3, 2010, doi: 10.1111/j.1365-2567.2009.03240.x.
P. Miossec, “IL-17 and Th17 cells in human inflammatory diseases,” Microbes Infect., vol. 11, no. 5, 2009, doi: 10.1016/j.micinf.2009.04.003.
Y. Qian, Z. Kang, C. Liu, and X. Li, “IL-17 signaling in host defense and inflammatory diseases,” Cell. Mol. Immunol., vol. 7, no. 5, pp. 328–333, 2010, doi: 10.1038/CMI.2010.27,.
H. I. Yoichiro Iwakura, “The IL-23/IL-17 axis in inflammation,” J. Clin. Invest., vol. 116, no. 5, 2006, doi: 10.1172/JCI28508.
R. J. H. Marc Veldhoen, “TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells,” Immunity, vol. 24, no. 2, 2006, doi: 10.1016/j.immuni.2006.01.001.
T. Korn, E. Bettelli, M. Oukka, and V. K. Kuchroo, “IL-17 and Th17 cells,” Annu. Rev. Immunol., vol. 27, pp. 485–517, 2009, doi: 10.1146/ANNUREV.IMMUNOL.021908.132710,.
R. A. Jeffrey A. Haspel, “Perfect timing: circadian rhythms, sleep, and immunity - an NIH workshop summary,” JCI Insight, vol. 5, no. 1, p. e131487, 2020, doi: 10.1172/jci.insight.131487.
A. A. Adam C. Silver, “Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells,” Brain. Behav. Immun., vol. 26, no. 3, 2012, doi: 10.1016/j.bbi.2011.10.001.
N. Cermakian et al., “Crosstalk between the circadian clock circuitry and the immune system,” Chronobiol. Int., vol. 30, no. 7, pp. 870–888, Aug. 2013, doi: 10.3109/07420528.2013.782315,.
J. S. Takahashi, “Transcriptional architecture of the mammalian circadian clock,” Nat. Rev. Genet., vol. 18, no. 3, pp. 164–179, Mar. 2017, doi: 10.1038/NRG.2016.150,.
F. D. Nicolas Preitner, “The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator,” Cell, vol. 110, no. 2, 2002, doi: 10.1016/s0092-8674(02)00825-5.
S. P. Trey K. Sato, “A functional genomics strategy reveals Rora as a component of the mammalian circadian clock,” Neuron, vol. 43, no. 4, pp. 527–37, 2004, doi: 10.1016/j.neuron.2004.07.018.
M. R. T. Jonathan M. Philpott, “Biochemical mechanisms of period control within the mammalian circadian clock,” Semin. Cell Dev. Biol., vol. 126, pp. 71–78, 2022, doi: 10.1016/j.semcdb.2021.04.012.
H. S. T. Ravasi, “An atlas of combinatorial transcriptional regulation in mouse and man,” Cell, vol. 140, no. 5, pp. 744–52, 2010, doi: 10.1016/j.cell.2010.01.044.
F. H. Akihiro Goriki, “A novel protein, CHRONO, functions as a core component of the mammalian circadian clock,” PLoS Biol., vol. 12, no. 4, 2014, doi: 10.1371/journal.pbio.1001839.
S. D. R. Arjun Sengupta, “Sleep restriction induced energy, methylation and lipogenesis metabolic switches in rat liver,” Int. J. Biochem. Cell Biol., vol. 93, pp. 129–135, 2017, doi: 10.1016/j.biocel.2017.08.014.
A. T. Mare Hallier, “The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb,” J. Biol. Chem., 1996, doi: 10.1074/jbc.271.19.11177.
T. M. Tetsuya Yamamoto, “Cross-talk between signal transducer and activator of transcription 3 and estrogen receptor signaling,” FEBS Lett., vol. 486, no. 2, 2000, doi: 10.1016/s0014-5793(00)02296-1.
M. X. Yongqiang Li, “Induction of CD4+ regulatory T cells by stimulation with Staphylococcal Enterotoxin C2 through different signaling pathways,” Biomed. Pharmacother., vol. 143, 2021, doi: 10.1016/j.biopha.2021.112204.
X. S. Xiang Xiao, “GITR subverts Foxp3(+) Tregs to boost Th9 immunity through regulation of histone acetylation,” Nat. Commun., vol. 6, no. 9, 2015, doi: 10.1038/ncomms9266.
D. Rudra et al., “Transcription factor Foxp3 and its protein partners form a complex regulatory network,” Nat. Immunol. 2012 1310, vol. 13, no. 10, pp. 1010–1019, Aug. 2012, doi: 10.1038/ni.2402.
M. Kanehisa, “Toward understanding the origin and evolution of cellular organisms,” Protein Sci., vol. 28, no. 11, pp. 1947–1951, 2019, doi: 10.1002/pro.3715.
E. D. Seth Carbon, “The Gene Ontology resource: enriching a GOld mine,” Nucleic Acids Res., vol. 49, no. D1, pp. D325–D334, 2021, doi: 10.1093/nar/gkaa1113.
H. H. Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, Duesbury M, Dumousseau M, Galeota E, Hinz U, Iannuccelli M, Jagannathan S, Jimenez R, Khadake J, Lagreid A, Licata L, Lovering RC, Meldal B, “The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases,” Nucleic Acids Res, 2014, doi: 10.1093/nar/gkt1115.
B. J. B. Chris Stark, “BioGRID: a general repository for interaction datasets,” Nucleic Acids Res., vol. 34, 2006, doi: 10.1093/nar/gkj109.
L. M. Bijay Jassal, “The reactome pathway knowledgebase,” Nucleic Acids Res., vol. 48, no. D1, pp. D498–D503, 2020, doi: 10.1093/nar/gkz1031.
E. C. Amnah Siddiqa, “Visualizing the regulatory role of Angiopoietin-like protein 8 (ANGPTL8) in glucose and lipid metabolic pathways,” Genomics, vol. 109, no. 5–6, pp. 408–418, 2017, doi: 10.1016/j.ygeno.2017.06.006.
D. D. Jérôme Jourquin, “GLAD4U: deriving and prioritizing gene lists from PubMed literature,” BMC Genomics, vol. 8, no. 8, p. 20, 2012, doi: 10.1186/1471-2164-13-S8-S20.
C. M. F. Caroline E. Sutton, “Loss of the molecular clock in myeloid cells exacerbates T cell-mediated CNS autoimmune disease,” Nat. Commun., vol. 8, p. 1923, 2017, doi: https://doi.org/10.1038/s41467-017-02111-0.
N. C. Nathalie Labrecque, “Circadian Clocks in the Immune System,” J. Biol. Rhythms, vol. 30, no. 4, p. 8, 2015, doi: 10.1177/0748730415577723.
F.-C. Chang, “A Dual Role for REV-ERB alpha in Th17 Cell Mediated Immune Response,” Univ. California, San Diego, 2016, [Online]. Available: https://escholarship.org/content/qt0hk9w237/qt0hk9w237_noSplash_c887d8d28515197a6d748ec7c9f1dce8.pdf?t=o8x15l
S. G. Teng F, Goc J, Zhou L, Chu C, Shah MA, Eberl G, “A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut,” Sci Immunol, 2019, doi: 10.1126/sciimmunol.aax1215.
S. B. Landgraf-Rauf K, Boeck A, Siemens D, Klucker E, Vogelsang V, Schmidt S, Kunze S, Weissenbacher C, Graessel A, Schmidt-Weber C, von Mutius E, Schedel M, “IRF-1 SNPs influence the risk for childhood allergic asthma: A critical role for pro-inflammatory immune regulation,” Pediatr Allergy Immunol, vol. 29, no. 1, pp. 34–41, 2018, doi: 10.1111/pai.12821.
C. T. Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, Wang L, Li N, Crestani E, Spielman S, Secor W, Biehl H, DiBenedetto N, Dong X, Umetsu DT, Bry L, Rachid R, “Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy,” Nat Med, vol. 25, no. 7, pp. 1164–1174, 2019, doi: 10.1038/s41591-019-0461-z.
Z. D. Tan W, Qiu Y, Chen N, Gao J, Liang J, Liu Y, “The intervention of intestinal Wnt/β-catenin pathway alters inflammation and disease severity of CIA,” Immunol Res, vol. 69, no. 4, pp. 323–333, 2021, doi: 10.1007/s12026-021-09190-8.
D. M. Subbanna M, Shivakumar V, Venugopal D, Narayanaswamy JC, Berk M, Varambally S, Venkatasubramanian G, “Impact of antipsychotic medication on IL-6/STAT3 signaling axis in peripheral blood mononuclear cells of drug-naive schizophrenia patients,” Psychiatry Clin Neurosci, vol. 74, no. 1, pp. 64–69, 2020, doi: 10.1111/pcn.12938.
Huai-Chia Chuang et al., “GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune disease,” Sci. Adv, 2018, doi: 10.1126/sciadv.aat5401.
M. H. George Assaf, “Coloured fuzzy Petri nets for modelling and analysing membrane systems,” BioSystems, vol. 212, p. 104592, 2022, doi: 10.1016/j.biosystems.2021.104592.
B. Aslam et al., “On the modelling and analysis of the regulatory network of dengue virus pathogenesis and clearance,” Comput. Biol. Chem., vol. 53, no. PB, pp. 277–291, 2014, doi: 10.1016/J.COMPBIOLCHEM.2014.10.003,.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 50sea

This work is licensed under a Creative Commons Attribution 4.0 International License.




















