Marek’s Disease and Its Outbreak in Asia: Python-Based Approach for Detection of Marek’s Virus
Keywords:
Marek’s Disease, Outbreak, PandemicAbstract
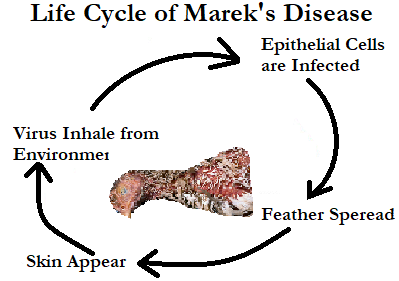
Marek's disease is an infectious disease that manifests in tumors of the nervous system and organs in chickens. Computer programming languages have enough potential to detect various viral diseases. An effort has been made to detect and evaluate the intensity of viruses. Despite the widespread use of effective vaccines designed to halt its spread, recent data reveal that their efficacy is declining as a result of the virus's adaptability. We analyzed 53 reports documenting 157 viral strains in Asian countries during the last decade of Marek's disease outbreaks and correlated meq sequences. The visceral variety of Marek's disease is the most common (18 out of 28 investigations), although there may be other, unrecognized brain alterations as well. Most commonly, MD causes tumors in the liver (16 out of 26 studies), however, other organs such as the spleen, kidney, heart, gizzard, skin, gut, lung, and sciatic nerve have also been affected. Using amino acid alignment, we found numerous point alterations in 28 strains that may be associated with its virulence. More research is needed on the virulence of the Marek strain, as well as the structural modifications to the Meq protein, and we recommend that this research take place in disease-endemic areas.
References
M. S. Parcells, V. Arumugaswami, J. T. Prigge, K. Pandya, and R. L. Dienglewicz, “Marek’s disease virus reactivation from latency: Changes in gene expression at the origin of replication,” Poult. Sci., vol. 82, no. 6, pp. 893–898, 2003, doi: 10.1093/ps/82.6.893.
S. H. S. Tai et al., “Expression of Marek’s Disease Virus Oncoprotein Meq During Infection in the Natural Host,” Virology, vol. 503, no. August 2016, pp. 103–113, 2017, doi: 10.1016/j.virol.2017.01.011.
Y. Yao and V. Nair, “Role of virus-encoded microRNAs in avian viral diseases,” Viruses, vol. 6, no. 3, pp. 1379–1394, 2014, doi: 10.3390/v6031379.
N. Chbab, A. Egerer, I. Veiga, K. W. Jarosinski, and N. Osterrieder, “Viral control of vTR expression is critical for efficient formation and dissemination of lymphoma induced by Marek’s disease virus (MDV),” Vet. Res., vol. 41, no. 5, 2010, doi: 10.1051/vetres/2010026.
V. Arumugaswami, P. M. Kumar, V. Konjufca, R. L. Dienglewicz, S. M. Reddy, and M. S. Parcells, “Latency of Marek’s Disease Virus (MDV) in a Reticuloendotheliosis Virus–transformed T-cell Line. I: Uptake and Structure of the Latent MDV Genome,” Avian Dis. Dig., vol. 4, no. 2, pp. e1–e1, 2009, doi: 10.1637/8861.1.
E. R. Tulman, C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish, “The Genome of a Very Virulent Marek’s Disease Virus,” J. Virol., vol. 74, no. 17, pp. 7980–7988, 2000, doi: 10.1128/jvi.74.17.7980-7988.2000.
C. Autexier and N. F. Lue, “The structure and function of telomerase reverse transcriptase,” Annu. Rev. Biochem., vol. 75, no. February 2006, pp. 493–517, 2006, doi: 10.1146/annurev.biochem.75.103004.142412.
A. Kheimar and B. B. Kaufer, “Epstein-Barr virus-encoded RNAs (EBERs) complement the loss of Herpesvirus telomerase RNA (vTR) in virus-induced tumor formation,” Sci. Rep., vol. 8, no. 1, pp. 1–8, 2018, doi: 10.1038/s41598-017-18638-7.
L. D. Bertzbach, A. Kheimar, F. A. Z. Ali, and B. B. Kaufer, “Viral Factors Involved in Marek’s Disease Virus (MDV) Pathogenesis,” Curr. Clin. Microbiol. Reports, vol. 5, no. 4, pp. 238–244, 2018, doi: 10.1007/s40588-018-0104-z.
N. Boodhoo, A. Gurung, S. Sharif, and S. Behboudi, “Marek’s disease in chickens: a review with focus on immunology,” Vet. Res., vol. 47, no. 1, pp. 1–19, 2016, doi: 10.1186/s13567-016-0404-3.
J. Schermuly, A. Greco, S. Härtle, N. Osterrieder, B. B. Kaufer, and B. Kaspers, “In vitro model for lytic replication, latency, and transformation of an oncogenic alphaherpesvirus,” Proc. Natl. Acad. Sci. U. S. A., vol. 112, no. 23, pp. 7279–7284, 2015, doi: 10.1073/pnas.1424420112.
A. C. Brown et al., “Interaction of MEQ protein and C-terminal-binding protein is critical for induction of lymphomas by Marek’s disease virus,” Proc. Natl. Acad. Sci. U. S. A., vol. 103, no. 6, pp. 1687–1692, 2006, doi: 10.1073/pnas.0507595103.
A. M. Levy et al., “Marek’s disease virus Meq transforms chicken cells via the v-Jun transcriptional cascade: A converging transforming pathway for avian oncoviruses,” Proc. Natl. Acad. Sci. U. S. A., vol. 102, no. 41, pp. 14831–14836, 2005, doi: 10.1073/pnas.0506849102.
B. Lupiani et al., “Marek’s disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication,” Proc. Natl. Acad. Sci. U. S. A., vol. 101, no. 32, pp. 11815–11820, 2004, doi: 10.1073/pnas.0404508101.
S. M. Reddy, B. Lupiani, I. M. Gimeno, R. F. Silva, L. F. Lee, and R. L. Witter, “Rescue of a pathogenic Marek’s disease virus with overlapping cosmid DNAs: Use of a pp38 mutant to validate the technology for the study of gene function,” Proc. Natl. Acad. Sci. U. S. A., vol. 99, no. 10, pp. 7054–7059, 2002, doi: 10.1073/pnas.092152699.
D. Jones, L. Lee, J. L. Liu, H. J. Rung, and J. K. Tillotson, “Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors,” Proc. Natl. Acad. Sci. U. S. A., vol. 89, no. 9, pp. 4042–4046, 1992, doi: 10.1073/pnas.89.9.4042.
R. F. Silva, J. R. Dunn, H. H. Cheng, and M. Niikura, “A MEQ-deleted marek’s disease virus cloned as a bacterial artificial chromosome is a highly efficacious vaccine,” Avian Dis., vol. 54, no. 2, pp. 862–869, 2010, doi: 10.1637/9048-090409-Reg.1.
K. A. Schat, B. J. L. Hooft Van Iddekinge, H. Boerrigter, P. H. O’Connell, and G. Koch, “Open reading frame L1 of Marek’s disease herpesvirus is not essential for in vitro and in vivo virus replication and establishment of latency,” J. Gen. Virol., vol. 79, no. 4, pp. 841–849, 1998, doi: 10.1099/0022-1317-79-4-841.
K. W. Jarosinski and K. A. Schat, “Multiple alternative splicing to exons II and III of viral interleukin-8 (vIL-8) in the Marek’s disease virus genome: The importance of vIL-8 exon I,” Virus Genes, vol. 34, no. 1, pp. 9–22, 2007, doi: 10.1007/s11262-006-0004-9.
A. Tahiri-Alaoui, L. P. Smith, L. Kgosana, L. J. Petherbridge, and V. Nair, “Identification of a neurovirulence factor from Marek’s disease virus,” Avian Dis., vol. 57, no. 2 SUPPL. 1, pp. 387–394, 2013, doi: 10.1637/10322-080912-Reg.1.
M. S. Parcells et al., “Marek’s Disease Virus (MDV) Encodes an Interleukin-8 Homolog (vIL-8): Characterization of the vIL-8 Protein and a vIL-8 Deletion Mutant MDV,” J. Virol., vol. 75, no. 11, pp. 5159–5173, 2001, doi: 10.1128/jvi.75.11.5159-5173.2001.
Z. Z. Cui, L. F. Lee, J. L. Liu, and H. J. Kung, “Structural analysis and transcriptional mapping of the Marek’s disease virus gene encoding pp38, an antigen associated with transformed cells,” J. Virol., vol. 65, no. 12, pp. 6509–6515, 1991, doi: 10.1128/jvi.65.12.6509-6515.1991.
H. J. Delecluse and W. Hammerschmidt, “Status of Marek’s disease virus in established lymphoma cell lines: herpesvirus integration is common,” J. Virol., vol. 67, no. 1, pp. 82–92, 1993, doi: 10.1128/jvi.67.1.82-92.1993.
Y. Hong and P. M. Coussens, “Identification of an immediate-early gene in the Marek’s disease virus long internal repeat region which encodes a unique 14-kilodalton polypeptide,” J. Virol., vol. 68, no. 6, pp. 3593–3603, 1994, doi: 10.1128/jvi.68.6.3593-3603.1994.
I. M. Gimeno, R. L. Witter, H. D. Hunt, S. M. Reddy, L. F. Lee, and R. F. Silva, “The pp38 Gene of Marek’s Disease Virus (MDV) Is Necessary for Cytolytic Infection of B Cells and Maintenance of the Transformed State but Not for Cytolytic Infection of the Feather Follicle Epithelium and Horizontal Spread of MDV,” J. Virol., vol. 79, no. 7, pp. 4545–4549, 2005, doi: 10.1128/jvi.79.7.4545-4549.2005.
X. Cui, L. F. Lee, W. M. Reed, H.-J. Kung, and S. M. Reddy, “Marek’s Disease Virus-Encoded vIL-8 Gene Is Involved in Early Cytolytic Infection but Dispensable for Establishment of Latency,” J. Virol., vol. 78, no. 9, pp. 4753–4760, 2004, doi: 10.1128/jvi.78.9.4753-4760.2004.
A. M. Levy et al., “Characterization of the Chromosomal Binding Sites andDimerization Partners of the Viral Oncoprotein Meq in Marek’sDisease Virus-Transformed TCells,” J. Virol., vol. 77, no. 23, pp. 12841–12851, 2003, doi: 10.1128/jvi.77.23.12841-12851.2003.
M. Teng et al., “Putative roles as oncogene or tumour suppressor of the midclustered microRNAs in Gallid alphaherpesvirus 2 (GaHV2) induced Marek’s disease lymphomagenesis,” J. Gen. Virol., vol. 98, no. 5, pp. 1097–1112, 2017, doi: 10.1099/jgv.0.000786.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 50SEA

This work is licensed under a Creative Commons Attribution 4.0 International License.




















