Cluster Analysis of COVID-19 Through Genome Sequences Using Python Bioinformatics Library
Keywords:
Covid-19, Genome sequences, nucleotide records, biomedical, pythonAbstract
Introduction and Importance of Study:
During the COVID-19 pandemic, mortality rates varied across different regions of the world. To better understand the virus's behavior, it's important to gain in-depth knowledge of the nucleotide records of the COVID-19 genomic sequence.
Novelty Statement:
In the study, researchers analyzed clusters of highly affected countries to find similar codons in countries with a similar effect of the virus through Python based library.
Material And Method:
Nucleotide records were extracted from the NCBI database in FASTA format. Further Python Bioinformatics library was used to form the clusters of each country using the K-means clustering technique.
Result and Discussion:
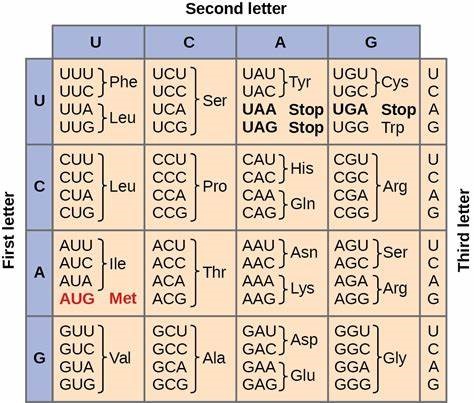
The study focuses on finding the similarities between the codons of amino acids in different countries that are affected in a similar way during COVID-19. For instance, China and the EU have a lower mortality rate and have Leucine, Methionine, Isoleucine, and Valine amino acids in common. On the other hand, countries like Pakistan and India have Leucine, Isoleucine, Valine, and Threonine in common and an average death rate. Moreover, Brazil and the US have a higher mortality rate and share similar codons such as Leucine, Glutamine, and Amber.
Concluding Remarks:
The study shows that countries affected by COVID-19 in a similar way share some common amino acids and their respective codons.
References
D. Cucinotta and M. Vanelli, “WHO Declares COVID-19 a Pandemic,” Acta Biomed., vol. 91, no. 1, pp. 157–160, 2020, doi: 10.23750/ABM.V91I1.9397.
A. N. Sajed and K. Amgain, “Coronavirus Disease (COVID-19) Outbreak and the Strategy for Prevention,” Eur. J. Med. Sci. , vol. 2, no. 1, pp. 1–3, Mar. 2020, doi: 10.46405/EJMS.V2I1.38.
F. Wu et al., “A new coronavirus associated with human respiratory disease in China,” Nat. 2020 5797798, vol. 579, no. 7798, pp. 265–269, Feb. 2020, doi: 10.1038/s41586-020-2008-3.
“COVID-19 cases | WHO COVID-19 dashboard.” Accessed: Mar. 03, 2024. [Online]. Available: https://data.who.int/dashboards/covid19/cases?n=c
S. Raskin, “Genetics of COVID-19,” J. Pediatr. (Rio. J)., vol. 97, no. 4, pp. 378–386, Jul. 2021, doi: 10.1016/J.JPED.2020.09.002.
I. Fricke-Galindo and R. Falfán-Valencia, “Genetics Insight for COVID-19 Susceptibility and Severity: A Review,” Front. Immunol., vol. 12, p. 622176, Apr. 2021, doi: 10.3389/FIMMU.2021.622176/BIBTEX.
P. D. Cristea, “Conversion of nucleotides sequences into genomic signals,” J. Cell. Mol. Med., vol. 6, no. 2, pp. 279–303, Apr. 2002, doi: 10.1111/J.1582-4934.2002.TB00196.X.
F. Castro-Chavez, “Most Used Codons per Amino Acid and per Genome in the Code of Man Compared to Other Organisms According to the Rotating Circular Genetic Code,” Neuroquantology, vol. 9, no. 4, pp. 747–767, 2011, doi: 10.14704/NQ.2011.9.4.500.
T. F. Clarke IV and P. L. Clark, “Rare Codons Cluster,” PLoS One, vol. 3, no. 10, p. e3412, Oct. 2008, doi: 10.1371/JOURNAL.PONE.0003412.
A. Baranova, H. Cao, S. Teng, K. P. Su, and F. Zhang, “Shared genetics and causal associations between COVID-19 and multiple sclerosis,” J. Med. Virol., vol. 95, no. 1, p. e28431, Jan. 2023, doi: 10.1002/JMV.28431.
K. S. te Paske, C. van Tienen, D. Dunk, D. van Pelt, and P. W. Smit, “SARS-CoV-2 transmission among health care workers, an outbreak investigation using whole-genome sequencing,” PLoS One, vol. 18, no. 3, p. e0283292, Mar. 2023, doi: 10.1371/JOURNAL.PONE.0283292.
H. C. Maltezou et al., “SARS-CoV-2 Infection in Healthcare Personnel With High-risk Occupational Exposure: Evaluation of 7-Day Exclusion From Work Policy,” Clin. Infect. Dis., vol. 71, no. 12, pp. 3182–3187, Dec. 2020, doi: 10.1093/CID/CIAA888.
J. Kim, S. Cheon, and I. Ahn, “NGS data vectorization, clustering, and finding key codons in SARS-CoV-2 variations,” BMC Bioinformatics, vol. 23, no. 1, pp. 1–24, Dec. 2022, doi: 10.1186/S12859-022-04718-7/TABLES/4.
B. Korber et al., “Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus,” Cell, vol. 182, no. 4, pp. 812-827.e19, Aug. 2020, doi: 10.1016/J.CELL.2020.06.043.
L. Zhang et al., “SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity,” Nat. Commun. 2020 111, vol. 11, no. 1, pp. 1–9, Nov. 2020, doi: 10.1038/s41467-020-19808-4.
L. Guruprasad, “Human SARS CoV-2 spike protein mutations,” Proteins Struct. Funct. Bioinforma., vol. 89, no. 5, pp. 569–576, May 2021, doi: 10.1002/PROT.26042.
D. Mercatelli and F. M. Giorgi, “Geographic and Genomic Distribution of SARS-CoV-2 Mutations,” Front. Microbiol., vol. 11, p. 555497, Jul. 2020, doi: 10.3389/FMICB.2020.01800/BIBTEX.
A. Umar, N. A. Mahoto, S. Bhatti, and S. Rathi, “Analysis of Covid-19 Genome Sequences based on Geo-Locations,” Pakistan J. Eng. Technol., vol. 4, no. 4, pp. 41–45, Dec. 2021, doi: 10.51846/VOL4ISS4PP41-45.
M. R. Islam et al., “Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity,” Sci. Reports 2020 101, vol. 10, no. 1, pp. 1–9, Aug. 2020, doi: 10.1038/s41598-020-70812-6.
N. Marascio et al., “Molecular Characterization and Cluster Analysis of SARS-CoV-2 Viral Isolates in Kahramanmaraş City, Turkey: The Delta VOC Wave within One Month,” Viruses 2023, Vol. 15, Page 802, vol. 15, no. 3, p. 802, Mar. 2023, doi: 10.3390/V15030802.
D. A. Benson, I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler, “GenBank,” Nucleic Acids Res., vol. 36, no. suppl_1, pp. D25–D30, Jan. 2008, doi: 10.1093/NAR/GKM929.
J. A. Botía et al., “An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks,” BMC Syst. Biol., vol. 11, no. 1, pp. 1–16, Apr. 2017, doi: 10.1186/S12918-017-0420-6/FIGURES/7.
H. Z. Girgis, “MeShClust v3.0: high-quality clustering of DNA sequences using the mean shift algorithm and alignment-free identity scores,” BMC Genomics, vol. 23, no. 1, pp. 1–16, Dec. 2022, doi: 10.1186/S12864-022-08619-0/FIGURES/3.
A. Melnyk et al., “From Alpha to Zeta: Identifying variants and subtypes of SARS-CoV-2 via clustering,” bioRxiv, p. 2021.08.26.457874, Aug. 2021, doi: 10.1101/2021.08.26.457874.
N. Shi, X. Liu, and Y. Guan, “Research on k-means clustering algorithm: An improved k-means clustering algorithm,” 3rd Int. Symp. Intell. Inf. Technol. Secur. Informatics, IITSI 2010, pp. 63–67, 2010, doi: 10.1109/IITSI.2010.74.
“Biopython · Biopython.” Accessed: Mar. 03, 2024. [Online]. Available: https://biopython.org/
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 50sea

This work is licensed under a Creative Commons Attribution 4.0 International License.




















