An Optimal Feature Extraction Technique for Glioma Tumor Detection from Brain MRI
Keywords:
Glioma Tumor, Classification, Feature Extraction, Dimensionality Reduction, Feature Selection, ,Linear Discriminant Analysis.Abstract
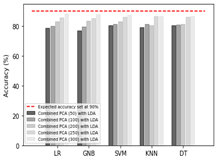
A brain tumor is defined by the uncontrolled proliferation of brain tissue cells, defying the typical cellular regulation mechanisms governing growth. The most significant challenge associated with brain tumors lies in their timely diagnosis and accurate stage determination. Accurate detection of tumors from MRI scans can not only assist doctors in their examination but also provide crucial information for appropriate and timely treatment decisions. In this paper, a comprehensive analysis is presented based on comparisons between state-of-the-art dimensionality reduction and classification algorithms. We used a dataset containing brain MRI scans, including both tumor and non-tumor cases, which was split into training and testing sets. After preprocessing the data, we implemented four feature extraction algorithms to obtain different sets of features. Consequently, these sets of features were used to train five classifiers to analyze the accuracy. Based on these results, optimal feature extraction and brain tumor classification technique is selected. The results indicate that the Linear Discriminant Analysis (LDA) technique extracted highly informative features, leading to an impressive accuracy of 92.84%. This highlights the effectiveness of LDA in significantly enhancing the performance of the brain tumor classification process, making it the prime choice for feature extraction that aligns seamlessly with the research's intuition. It has higher accuracy with all the classifiers.
References
“Brain Tumor MRI Dataset.” https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset (accessed Nov. 02, 2023).
“Brain Tumor Classification (MRI).” https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri (accessed Nov. 02, 2023).
B. Ilhan, P. Guneri, and P. Wilder-Smith, “The contribution of artificial intelligence to reducing the diagnostic delay in oral cancer,” Oral Oncol., vol. 116, p. 105254, May 2021, doi: 10.1016/J.ORALONCOLOGY.2021.105254.
A. Osman and B. Alsabbagh, “Image-classification for Brain Tumor using Pre-trained Convolutional Neural Network”.
M. Westphal and K. Lamszus, “The neurobiology of gliomas: from cell biology to the development of therapeutic approaches,” Nat. Rev. Neurosci. 2011 129, vol. 12, no. 9, pp. 495–508, Aug. 2011, doi: 10.1038/nrn3060.
L. R. S. Charfi, R. Lahmyed, “A novel approach for brain tumor detection using neural network,” Int. J. Res. Eng. Technol., vol. 2, no. 7, pp. 93–104, 2014.
B. Ural, “A Computer-Based Brain Tumor Detection Approach with Advanced Image Processing and Probabilistic Neural Network Methods,” J. Med. Biol. Eng., vol. 38, no. 6, pp. 867–879, Dec. 2018, doi: 10.1007/S40846-017-0353-Y/METRICS.
K. Sudharani, T. C. Sarma, and K. S. Prasad, “Advanced Morphological Technique for Automatic Brain Tumor Detection and Evaluation of Statistical Parameters,” Procedia Technol., vol. 24, pp. 1374–1387, Jan. 2016, doi: 10.1016/J.PROTCY.2016.05.153.
A. S. A. and P. Augustine, “Efficient brain tumor classification using PCA and SVM,” Int. J. Res. Eng. IT Soc. Sci., vol. 7, pp. 1–7, 2017.
“Deep learning for brain tumor classification.” https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10137/1/Deep-learning-for-brain-tumor-classification/10.1117/12.2254195.full (accessed Nov. 02, 2023).
I. Guyon and A. Elisseefl, “An introduction to feature extraction,” Stud. Fuzziness Soft Comput., vol. 207, pp. 1–25, 2006, doi: 10.1007/978-3-540-35488-8_1/COVER.
R. Bro and A. K. Smilde, “Principal component analysis,” Anal. Methods, vol. 6, no. 9, pp. 2812–2831, Apr. 2014, doi: 10.1039/C3AY41907J.
P. Xanthopoulos, P. M. Pardalos, and T. B. Trafalis, “Linear Discriminant Analysis,” pp. 27–33, 2013, doi: 10.1007/978-1-4419-9878-1_4.
J. V. Stone, “Independent component analysis: an introduction,” Trends Cogn. Sci., vol. 6, no. 2, pp. 59–64, 2002.
N. Khosla, “Dimensionality reduction using factor analysis,” Griffith Univ. Aust., 2004.
J. R. Lambert and E. Perumal, “Optimal Feature Selection Methods for Chronic Kidney Disease Classification using Intelligent Optimization Algorithms,” Recent Adv. Comput. Sci. Commun., vol. 14, no. 9, pp. 2886–2898, Aug. 2020, doi: 10.2174/2666255813999200818131835.
H. Kamel, D. Abdulah, and J. M. Al-Tuwaijari, “Cancer Classification Using Gaussian Naive Bayes Algorithm,” Proc. 5th Int. Eng. Conf. IEC 2019, pp. 165–170, Jun. 2019, doi: 10.1109/IEC47844.2019.8950650.
W. S. Noble, “What is a support vector machine?,” Nat. Biotechnol., vol. 24, no. 12, pp. 1565–1567, Dec. 2006, doi: 10.1038/NBT1206-1565.
Y. Liao and V. R. Vemuri, “Use of K-Nearest Neighbor classifier for intrusion detection,” Comput. Secur., vol. 21, no. 5, pp. 439–448, Oct. 2002, doi: 10.1016/S0167-4048(02)00514-X.
A. J. Myles, R. N. Feudale, Y. Liu, N. A. Woody, and S. D. Brown, “An introduction to decision tree modeling,” J. Chemom., vol. 18, no. 6, pp. 275–285, Jun. 2004, doi: 10.1002/CEM.873.
M. Hossin and Sulaiman, “A REVIEW ON EVALUATION METRICS FOR DATA CLASSIFICATION EVALUATIONS,” Int. J. Data Min. Knowl. Manag. Process, vol. 5, no. 2, 2015, doi: 10.5121/ijdkp.2015.5201.
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 50SEA

This work is licensed under a Creative Commons Attribution 4.0 International License.




















