An Artificial Intelligence Vision Transformer Model for Classification of Bacterial Colony
Keywords:
Classification, deep learning algorithms, E. coli, Salmonella, Vision TransformerAbstract
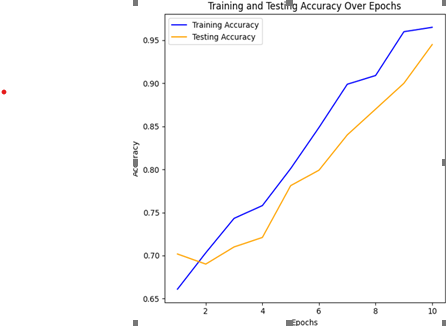
The application of AI and machine learning, particularly the vision transformer method, in bacterial detection presents a promising solution to overcome limitations of traditional methods, offering faster and more accurate detection of disease-causing bacteria like E. coli and salmonella in water, crucial for human survival, with ongoing research to further assess its effectiveness in microbiology. This research introduces a revolutionary positional self-attention transformer model for the classification of bacterial colonies. Leveraging the proven success of transformer architectures in various domains, we enhanced the model's performance by integrating a positional self-attention mechanism. We presented a novel approach for bacterial colony classification utilizing a positional self-attention transformer model. This allows the model to effectively capture spatial relationships and patterns within bacterial colonies, contributing to highly accurate classification results. We trained the model on a substantial dataset of bacterial images, which ensures its robustness and generalization to diverse colony types. The proposed model adeptly captured the spatial relationships and sequential patterns inherent in bacterial colony images, allowing for more accurate and robust classification. The proposed model demonstrated remarkable performance, achieving an accuracy of 98.50% in the classification of bacterial colonies. This novel approach surpasses traditional methods by effectively capturing intricate spatial relationships within microbial structures, offering unprecedented accuracy in discerning subtle morphological variations. The model's adaptability to diverse colony shapes and arrangements marks a significant advancement, promising to redefine the landscape of bacterial colony classification through the lens of state-of-the-art deep learning techniques. The high classification accuracy attained by the model, suggests its potential for practical applications in the early diagnosis of infectious diseases and the development of targeted treatments. The findings of this study underscore the effectiveness of incorporating positional self-attention in transformer models for image-based classification tasks, particularly in the domain of bacterial colony analysis.
References
“Determination of E. Coli Bacteria in Drinking Waters Using Image Processing Techniques,” 2019, [Online]. Available: https://dergipark.org.tr/tr/pub/cukurovaummfd/issue/49748/638164
M. Poladia, P. Fakatkar, S. Hatture, S. S. Rathod, and S. Kuruwa, “Detection and analysis of waterborne bacterial colonies using image processing and smartphones,” 2015 Int. Conf. Smart Technol. Manag. Comput. Commun. Control. Energy Mater. ICSTM 2015 - Proc., pp. 159–164, Aug. 2015, doi: 10.1109/ICSTM.2015.7225406.
C. Zhang, G. Lin, F. Liu, J. Guo, Q. Wu, and R. Yao, “Pyramid graph networks with connection attentions for region-based one-shot semantic segmentation,” Proc. IEEE Int. Conf. Comput. Vis., vol. 2019-October, pp. 9586–9594, Oct. 2019, doi: 10.1109/ICCV.2019.00968.
F. F. Farrell, M. Gralka, O. Hallatschek, and B. Waclaw, “Mechanical interactions in bacterial colonies and the surfing probability of beneficial mutations,” bioRxiv, p. 100099, Jan. 2017, doi: 10.1101/100099.
S. İlkin, T. H. Gençtürk, F. Kaya Gülağız, H. Özcan, M. A. Altuncu, and S. Şahin, “hybSVM: Bacterial colony optimization algorithm based SVM for malignant melanoma detection,” Eng. Sci. Technol. an Int. J., vol. 24, no. 5, pp. 1059–1071, Oct. 2021, doi: 10.1016/J.JESTCH.2021.02.002.
C. Shorten and T. M. Khoshgoftaar, “A survey on Image Data Augmentation for Deep Learning,” J. Big Data, vol. 6, no. 1, pp. 1–48, Dec. 2019, doi: 10.1186/S40537-019-0197-0/FIGURES/33.
H. Alshammari, K. Gasmi, I. Ben Ltaifa, M. Krichen, L. Ben Ammar, and M. A. Mahmood, “Olive Disease Classification Based on Vision Transformer and CNN Models,” Comput. Intell. Neurosci., vol. 2022, 2022, doi: 10.1155/2022/3998193.
H. Yanik, A. Hilmi Kaloğlu, and E. Değirmenci, “Detection of Escherichia Coli Bacteria in Water Using Deep Learning: A Faster R-CNN Approach,” Teh. Glas., vol. 14, no. 3, pp. 273–280, Sep. 2020, doi: 10.31803/TG-20200524225359.
S. Wu, Y. Sun, and H. Huang, “Multi-granularity Feature Extraction Based on Vision Transformer for Tomato Leaf Disease Recognition,” 2021 3rd Int. Acad. Exch. Conf. Sci. Technol. Innov. IAECST 2021, pp. 387–390, 2021, doi: 10.1109/IAECST54258.2021.9695688.
A. Dosovitskiy et al., “An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale,” ICLR 2021 - 9th Int. Conf. Learn. Represent., Oct. 2020, Accessed: Feb. 21, 2024. [Online]. Available: https://arxiv.org/abs/2010.11929v2
J. W. Yuhui Yuan, Xiaokang Chen, Xilin Chen, “Segmentation Transformer: Object-Contextual Representations for Semantic Segmentation”, [Online]. Available: https://arxiv.org/abs/1909.11065
H. Wang et al., “Early detection and classification of live bacteria using time-lapse coherent imaging and deep learning,” Light Sci. Appl. 2020 91, vol. 9, no. 1, pp. 1–17, Jul. 2020, doi: 10.1038/s41377-020-00358-9.
P. T. Su, C. T. Liao, J. R. Roan, S. H. Wang, A. Chiou, and W. J. Syu, “Bacterial Colony from Two-Dimensional Division to Three-Dimensional Development,” PLoS One, vol. 7, no. 11, p. e48098, Nov. 2012, doi: 10.1371/JOURNAL.PONE.0048098.
M. Bedrossian et al., “A machine learning algorithm for identifying and tracking bacteria in three dimensions using Digital Holographic Microscopy,” AIMS Biophys. 2018 136, vol. 5, no. 1, pp. 36–49, 2018, doi: 10.3934/BIOPHY.2018.1.36.
B. Zieliński, A. Plichta, K. Misztal, P. Spurek, M. Brzychczy-Włoch, and D. Ochońska, “Deep learning approach to bacterial colony classification,” PLoS One, vol. 12, no. 9, p. e0184554, Sep. 2017, doi: 10.1371/JOURNAL.PONE.0184554.
H. Liu, C. A. Whitehouse, and B. Li, “Presence and Persistence of Salmonella in Water: The Impact on Microbial Quality of Water and Food Safety,” Front. Public Heal., vol. 6, p. 366505, May 2018, doi: 10.3389/FPUBH.2018.00159/BIBTEX.
A. Fiannaca et al., “Deep learning models for bacteria taxonomic classification of metagenomic data,” BMC Bioinformatics, vol. 19, no. 7, pp. 61–76, Jul. 2018, doi: 10.1186/S12859-018-2182-6/TABLES/5.
K. P. Ferentinos, “Deep learning models for plant disease detection and diagnosis,” Comput. Electron. Agric., vol. 145, pp. 311–318, Feb. 2018, doi: 10.1016/J.COMPAG.2018.01.009.
M. M. Saritas and A. Yasar, “Performance Analysis of ANN and Naive Bayes Classification Algorithm for Data Classification,” Int. J. Intell. Syst. Appl. Eng., vol. 7, no. 2, pp. 88–91, Jun. 2019, doi: 10.18201//ijisae.2019252786.
A. Vaswani et al., “Attention Is All You Need,” Adv. Neural Inf. Process. Syst., vol. 2017-December, pp. 5999–6009, Jun. 2017, Accessed: Oct. 01, 2023. [Online]. Available: https://arxiv.org/abs/1706.03762v7
L. C. Chen, G. Papandreou, I. Kokkinos, K. Murphy, and A. L. Yuille, “DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 40, no. 4, pp. 834–848, Apr. 2018, doi: 10.1109/TPAMI.2017.2699184.
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 50SEA

This work is licensed under a Creative Commons Attribution 4.0 International License.




















