A Hybrid Approach to Fine-Grained Butterfly and Moth Classification Using Deep Features and Rhombus-Based HOG Descriptor
Keywords:
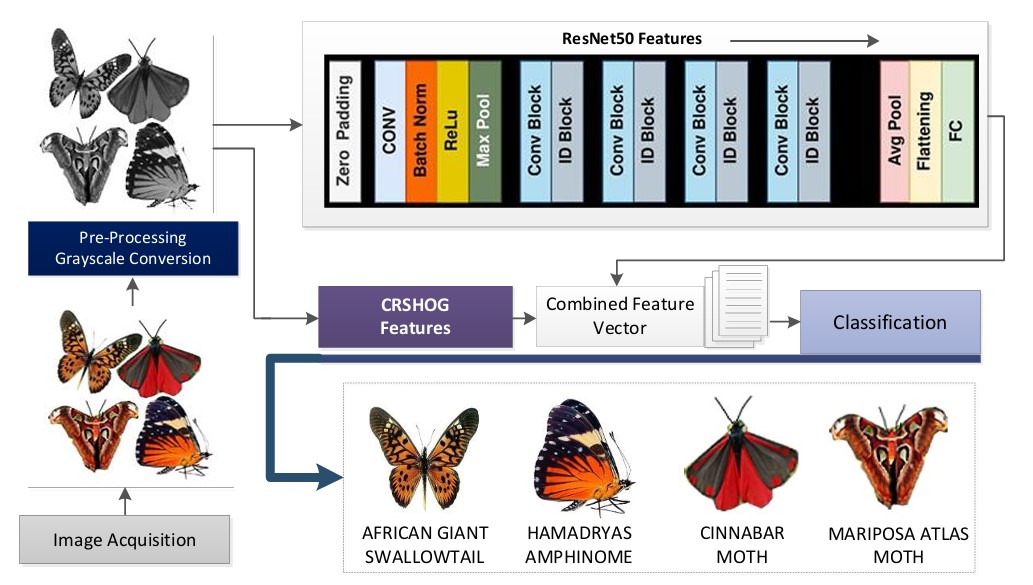
Feature Extraction, Computer Vision, Deep Learning, CLAHE, Corner Rhombus Shape HOG (CRSHOG), ResNet-50Abstract
Butterfly and moth species are crucial for ecosystems as pollinators, pests, and biodiversity indicators, therefore necessitating their precise automated classification for extensive monitoring, conservation initiatives, and agricultural pest control. Nonetheless, considerable obstacles emerge from inter- and intra-species variety in wing coloration, patterns, posture, and the effects of lighting and background circumstances on pictures. This study presents a comprehensive framework that enhances feature representation via a dual-phase methodology. Initially, pictures undergo preprocessing by Contrast-Limited Adaptive Histogram Equalization (CLAHE) to augment distinguishing features. Subsequently, elevated semantic features are derived using a ResNet50 backbone pre-trained on ImageNet, with a baseline accuracy of 92%. A unique Corner Rhombus Shape HOG (CRSHOG) descriptor is suggested to accurately capture detailed geometric and textural wing properties, utilizing rhombus-based grid sampling and gradient orientation encoding. These complementary deep and handcrafted features are carefully integrated to form a hybrid representation, improving resilience to cluttered backdrops and position changes. The integrated feature set is assessed using several classifiers, with an Ensemble Subspace KNN model attaining the greatest classification accuracy of 94.6% on the Butterfly and Moth Image dataset, exceeding traditional CNN (Convolutional Neural Network)-only and HOG-based methods. These findings highlight the benefits of combining domain-specific shape descriptors with deep-learning features to enhance fine-grained insect categorization. Moreover, depending exclusively on standard RGB photos facilitates practical implementation on mobile and aerial platforms for real-time biodiversity surveillance and pest management. Future endeavors will concentrate on expanding this hybrid feature technique to encompass live video tracking and open-set species detection in uncontrolled settings.
References
A. Hoskins, “Butterflies of the world,” London NH (Reed New Holl. Publ., 2015, [Online]. Available: https://archive.org/details/butterfliesofwor0000hosk
C. L. Boggs, W. B. Watt, and P. R. Ehrlich, “Butterflies Ecology and Evolution Taking Flight,” Univ. Chicago Press, 2003, doi: 10.7208/CHICAGO/9780226063195.001.0001.
A. S. Almryad and H. Kutucu, “Automatic identification for field butterflies by convolutional neural networks,” Eng. Sci. Technol. an Int. J., vol. 23, no. 1, pp. 189–195, 2020, [Online]. Available: https://www.sciencedirect.com/science/article/pii/S2215098619326011?via%3Dihub
J. Boulent, S. Foucher, J. Théau, and P.-L. St-Charles, “Convolutional Neural Networks for the Automatic Identification of Plant Diseases,” Front. Plant Sci., vol. 10, p. 941, Jul. 2019, doi: 10.3389/FPLS.2019.00941.
A. P. Wibowo and D. R. I. M. Setiadi, “Performance Analysis of Deep Learning Models for Sweet Potato Image Recognition,” 2022 Int. Semin. Appl. Technol. Inf. Commun. Technol. 4.0 Smart Ecosyst. A New W. Doing Digit. Business, iSemantic 2022, pp. 101–106, 2022, doi: 10.1109/ISEMANTIC55962.2022.9920423.
A. Ghosh, A. Sufian, F. Sultana, A. Chakrabarti, and D. De, “Fundamental Concepts of Convolutional Neural Network,” Intell. Syst. Ref. Libr., vol. 172, pp. 519–567, 2020, doi: 10.1007/978-3-030-32644-9_36.
M. A. S. A. Fathimathul Rajeena P. P., Rasha Orban, Kogilavani Shanmuga Vadivel, Malliga Subramanian, Suresh Muthusamy, Diaa Salam Abd Elminaam, Ayman Nabil, Laith Abulaigh, Mohsen Ahmadi, “A Novel Method for the Classification of Butterfly Species Using Pre-Trained CNN Models,” Electronics, vol. 11, no. 13, p. 2016, 2022, doi: https://doi.org/10.3390/electronics11132016.
R. Rashmi, U. Snekhalatha, A. L. Salvador, and A. N. J. Raj, “Facial emotion detection using thermal and visual images based on deep learning techniques,” Imaging Sci. J., vol. 72, no. 2, pp. 153–166, Feb. 2024, doi: 10.1080/13682199.2023.2199504;SUBPAGE:STRING:ACCESS.
H. A. Khan and S. M. Adnan, “A Novel Fusion Technique for Early Detection of Alopecia Areata Using ResNet-50 and CRSHOG,” IEEE Access, vol. 12, pp. 139912–139922, 2024, doi: 10.1109/ACCESS.2024.3461324.
A. S. R. M. A. Erick A. Adjé, Gilles Delmaire, “A Novel Statistical Framework for Butterfly Species Recognition Using Raw Spatio-Hyperspectral Images,” Res. Sq., 2024, [Online]. Available: https://www.researchsquare.com/article/rs-5240841/v1
Jayme Garcia Arnal Barbedo, “Detecting and Classifying Pests in Crops Using Proximal Images and Machine Learning: A Review,” AI, vol. 1, no. 2, pp. 312–328, 2020, doi: https://doi.org/10.3390/ai1020021.
Y. G. Xin Chen, Bin Wang, “Separated Fan-Beam Projection with Gaussian Convolution for Invariant and Robust Butterfly Image Retrieval.,” Pattern Recognit., vol. 147, p. 110083, 2024, doi: https://doi.org/10.1016/j.patcog.2023.110083.
Y. Rong, H. Su, W. Zhang, and Z. Li, “Fine-Grained Complex Image Classification Method Based on Butterfly Images,” Proc. 2022 6th Asian Conf. Artif. Intell. Technol. ACAIT 2022, 2022, doi: 10.1109/ACAIT56212.2022.10137872.
J. U. Hilmi Syamsudin, Saidatul Khalidah, “Lepidoptera Classification Using Convolutional Neural Network EfficientNet-B0,” Indones. J. Artifical Intelligance Data Min., vol. 7, no. 1, 2024, [Online]. Available: https://ejournal.uin-suska.ac.id/index.php/IJAIDM/article/view/24586
X. Jia, X. Tan, G. Jin, and R. O. Sinnott, “Lepidoptera Classification through Deep Learning,” 2020 IEEE Asia-Pacific Conf. Comput. Sci. Data Eng. CSDE 2020, Dec. 2020, doi: 10.1109/CSDE50874.2020.9411382.
S. S. Abdul Fadlil Abdul Fadlil, Ainin Maftukhah, “Butterfly Image Identification Using Multilevel Thresholding Segmentasi and Convolution Neural Network Classification with Alexnet Architecture,” Int. J. Comput. Digit. Syst., vol. 15, no. 1, pp. 1775–1785, 2024, doi: 10.12785/ijcds/1501125.
Harish Trio Adityawan, Omar Farroq, Stefanus Santosa, Hussain Md Mehedul Islam, Md Kamruzzaman Sarker, “Butterflies Recognition using Enhanced Transfer Learning and Data Augmentation,” J. Comput. Theor. Appl., vol. 1, no. 2, 2023, [Online]. Available: http://publikasi.dinus.ac.id/index.php/jcta/article/view/9443
Hari Theivaprakasham, “Identification of Indian butterflies using Deep Convolutional Neural Network,” J. Asia. Pac. Entomol., vol. 24, no. 1, pp. 329–340, 2021, doi: https://doi.org/10.1016/j.aspen.2020.11.015.
T. Xi, “Multiple butterfly recognition based on deep residual learning and image analysis,” Entomol. Res., vol. 52, no. 1, pp. 44–53, 2022, [Online]. Available: https://onlinelibrary.wiley.com/doi/epdf/10.1111/1748-5967.12564
T. Surabhi, B. Sachin, and C. Advait, “Deep Convolutional Neural Networks for Automated Butterfly Species Recognition and Classification,” Proc. 5th Int. Conf. Inven. Res. Comput. Appl. ICIRCA 2023, pp. 778–783, 2023, doi: 10.1109/ICIRCA57980.2023.10220696.
Fengwei Liu, “Butterfly and Moth Image Recognition Based on Residual Neural Network,” Int. Conf. Data Anal. Mach. Learn., pp. 211–217, 2023, doi: 10.5220/0012798500003885.
S. Y. and X. F. Ruoyan Zhao, Cuixia Li, “Butterfly Recognition Based on Faster R-CNN,” J. Phys. Conf. Ser., vol. 1176, no. 3, 2019, doi: 10.1088/1742-6596/1176/3/032048.
N. N. Kamaron Arzar, N. Sabri, N. F. Mohd Johari, A. Amilah Shari, M. R. Mohd Noordin, and S. Ibrahim, “Butterfly Species Identification Using Convolutional Neural Network (CNN),” 2019 IEEE Int. Conf. Autom. Control Intell. Syst. I2CACIS 2019 - Proc., pp. 221–224, Jun. 2019, doi: 10.1109/I2CACIS.2019.8825031.
B. W. Kunkun Zhang, Xin Chen, “MdeBEIA: Multi-task deep leaning for butterfly ecological image analysis,” Digit. Signal Process., vol. 162, p. 105168, 2025, [Online]. Available: https://www.sciencedirect.com/science/article/abs/pii/S1051200425001903?via%3Dihub
J. Kaur, S. Rani, and G. Singh, “Evolving Patterns: A CNN Approach to Butterfly Species Recognition,” 8th Int. Conf. Electron. Commun. Aerosp. Technol. ICECA 2024 - Proc., pp. 1000–1005, 2024, doi: 10.1109/ICECA63461.2024.10800980.
S. A. Samruddhi N Hegde, Manjesh R, P Sindhura, Reema Rodrigues, “Identification of butterfly speciecs using Vgg-16,” Int. Res. J. Mod. Eng. Technol. Sci., vol. 2, no. 5, 2020, [Online]. Available: https://www.irjmets.com/uploadedfiles/paper/volume2/issue_5_may_2020/1088/1628083020.pdf
Gerry, “Butterfly & Moths Image Classification 100 species,” Kaggle, 2023, [Online]. Available: https://www.kaggle.com/datasets/gpiosenka/butterfly-images40-species
Y. Yoshimi et al., “Image preprocessing with contrast-limited adaptive histogram equalization improves the segmentation performance of deep learning for the articular disk of the temporomandibular joint on magnetic resonance images,” Oral Surg. Oral Med. Oral Pathol. Oral Radiol., vol. 138, no. 1, pp. 128–141, Jul. 2024, doi: 10.1016/J.OOOO.2023.01.016,.
M. P. Thi Phuoc Hanh Nguyen, Zinan Cai, Khanh Nguyen, Sokuntheariddh Keth, Ningyuan Shen, “Pre-processing Image using Brightening, CLAHE and RETINEX,” arXiv:2003.10822, 2020, doi: https://doi.org/10.48550/arXiv.2003.10822.
R. Bhuria, “Feature Extraction and Classification Performance of Fine-Tuned ResNet-50 in Plant Species Recognition,” 2024 2nd Int. Conf. Adv. Comput. Commun. Inf. Technol. ICAICCIT 2024, pp. 574–579, 2024, doi: 10.1109/ICAICCIT64383.2024.10912176.
D. L. Bin Li, “Facial expression recognition via ResNet-50,” Int. J. Cogn. Comput. Eng., vol. 2, pp. 57–64, 2021, [Online]. Available: https://www.sciencedirect.com/science/article/pii/S2666307421000073?via%3Dihub
Suhita Ray, “Disease Classification within Dermascopic Images Using features extracted by ResNet50 and classification through Deep Forest,” arXiv:1807.05711, 2018, doi: https://doi.org/10.48550/arXiv.1807.05711.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 50sea

This work is licensed under a Creative Commons Attribution 4.0 International License.




















